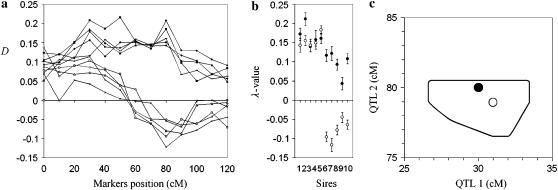
Figure 4.—
Analysis with multiple-linked QTL. Simulated were 10 families heterozygous for two linked QTL, 5 in coupling and 5 in repulsion phase. Thirteen markers were evenly spaced on a chromosome of length 120 cM. QTL 1 and QTL 2 were simulated in positions 30 and 80 cM, respectively. The allele substitution effect at both QTL in all 10 families was d/σ = 0.3. Alleles at QTL 1 and QTL 2 that came from dams were simulated as independent cases. The number of daughters per family was 2000; the proportion of total population selected to each tail was 0.10. (a) D-values for all families and markers. Points corresponding to a given family are connected by a line. (b) λ-Values and their standard errors in 500 jackknifes for every family. Clear separation is observed between the first five sires (QTL in coupling phase) and the last five sires (QTL in repulsion phase). (c) Simulated (solid circle) and estimated (open circle) positions of QTL. The curve encloses the area where the position of QTL was estimated in ≥90% of 500 jackknifes {included points with integer coordinates (x, y) such that in ≥5 jackknifes, estimated QTL positions belonged in the interval (x ± 0.5, y ± 0.5 cM).