TABLE 1.
Effect of number of markers (M) under the FPD on the confidence interval (C.I.) of QTL location, comparisonwise error rate (P-value), and statistical power, according to the test for significance and standardized allele substitution effect at the QTL (d/σ), using simulated data
| C.I.
|
P-value
|
Power
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| d/σ | M | Δ | SD | Σ(λf)2 | max|λ| | max-χ2 | 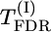 |
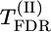 |
Σ(λf)2 (%) | max-χ2 (%) |
| 0.2 | 25 | 4.1 | 3.1 | 0.003 | 0.053 | 0.007 | 0.015 | 0.074 | 99 | 56 |
| 13 | 5.1 | 3.3 | 0.002 | 0.030 | 0.008 | 0.018 | 0.076 | 99 | 59 | |
| 7 | 6.4 | 3.6 | 0.004 | 0.049 | 0.006 | 0.016 | 0.061 | 98 | 64 | |
| 0.15 | 25 | 4.9 | 5.2 | 0.008 | 0.104 | 0.056 | 0.067 | 0.250 | 92 | 27 |
| 13 | 6.3 | 5.2 | 0.021 | 0.071 | 0.126 | 0.112 | 0.260 | 90 | 24 | |
| 7 | 7.8 | 5.9 | 0.021 | 0.098 | 0.130 | 0.101 | 0.299 | 89 | 28 | |
Tests of significance: Σ(λf)2, max|λ|, max-χ2, 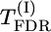 , and
, and 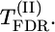 See text for details. Power was calculated at P-value = 5%. Values Δ and SD characterize the center and size of the confidence interval obtained in jackknife iterations (see text). Parameters of the simulations: chromosome length 120 cM. A single QTL was situated in position 40 cM. Number of families, F = 10 (5 families, sire heterozygous at the QTL; 5 families, sire homozygous at the QTL); number of daughters per family, N = 2000; proportion of the population selected to each tail, 0.10; number of subpools per tail, S = 4. Values are the mean based on 10 simulation data sets; for every data set, 500 permutations and 100 jackknife iterations were made.
See text for details. Power was calculated at P-value = 5%. Values Δ and SD characterize the center and size of the confidence interval obtained in jackknife iterations (see text). Parameters of the simulations: chromosome length 120 cM. A single QTL was situated in position 40 cM. Number of families, F = 10 (5 families, sire heterozygous at the QTL; 5 families, sire homozygous at the QTL); number of daughters per family, N = 2000; proportion of the population selected to each tail, 0.10; number of subpools per tail, S = 4. Values are the mean based on 10 simulation data sets; for every data set, 500 permutations and 100 jackknife iterations were made.
