Abstract
Plant O-methyltransferases (OMTs) play important roles in secondary metabolism. Two clusters of genes coding for caffeic acid OMT (COMT) have been identified in the apple genome. Three genes from one cluster and two genes from another cluster were isolated. These five genes encoding COMT, designated Mdomt1–Mdomt5 (GenBank accession nos. DQ886018–DQ886022), were distinguished by a (CT)n microsatellite in the 5′-UTR and two transposon-like sequences present in the promoter region and intron 1, respectively. The transposon-like sequence in intron 1 unambiguously traced the five Mdomt genes in the apple to a common ancestor. The ancestor must have undergone an initial duplication generating two progenitors, and this was followed by further duplication of these progenitors resulting in the two clusters identified in this study. The distal regions of the transposon-like sequences in promoter regions of Mdomt genes are capable of forming palindromic hairpin-like structures. The hairpin formation is likely responsible for nucleotide sequence differences observed in the promoter regions of these genes as it plays a destabilizing role in eukaryotic chromosomes. In addition, the possible mechanism of amplification of Mdomt genes in the apple genome is also discussed.
SECONDARY metabolites such as lignin, flavonoids, anthocyanins, suberin, and isoflavonoids are abundant in plants and they play various roles in plant growth and development as well as in plant interactions with the environment, including defense responses against microorganisms and herbivores (Schwab 2003). Plant O-methyltransferases (OMTs) play important roles in secondary metabolism, and many OMTs have been identified in plants. To date, the most thoroughly studied OMTs are caffeic acid OMT (COMT) and caffeoyl CoA OMT (CCOMT). While both are involved in lignin biosynthesis, COMT methylates caffeic acid/5-hydroxyferulic acid, whereas, CCOMT methylates CoA ester (Ye and Varner 1995; Li et al. 1997; Inoue et al. 1998; Maury et al. 1999). Flavonoid and isoflavone OMTs involved in the biosynthesis of phytoalexins have also been identified (Christensen et al. 1998; He et al. 1998). More recently, various OMTs involved in the biosynthetic pathways of floral scent components have been identified and characterized. For example, S-adenosyl-l-methionine (iso) eugenol OMT (IEMT), which catalyzes eugenol and isoeugenol to form volatile methyleugenol and isomethyleugenol, was isolated from Clarkia breweri (Wang et al. 1997). Eugenol OMT (EOMT) and chavicol OMT (CVOMT), which convert eugenol and chavicol to methyleugenol and methylchavicol, respectively, have been identified in Ocimun basilicum (Gang et al. 2002). Also, two orcinol OMTs (OOMT), OOMT1 and OOMT2, have been isolated from Rosa hybrida, and these efficiently methylate orcinol to produce 3,5-dimethoxytoluene (Lavid et al. 2002). Moreover, two OMTs involved in scent biosynthesis, RcOMT1 and RcOMT2, have also been isolated from R. chinensis (Wu et al. 2003).
Although some OMT members have multiple functions (Li et al. 1997; Gauthier et al. 1998), most generally exhibit high substrate specificity (Ibrahim et al. 1998). Recent studies have demonstrated that this substrate specificity can be altered by mutations of either a single or a few amino acid(s), thus suggesting that functionally distinct genes encoding OMTs may have evolved from a common ancestral gene via duplication and mutation (Gang 2005). For example, although two genes encoding EOMT1 and CVOMT1 from O. basilicum share 90% amino acid identity, a single-amino-acid difference is responsible for the substrate discrimination between CVOMT1 and EOMT1 (Gang et al. 2002). This key amino acid difference arose from a C–T transition, which is the most common form of observed DNA mutation. Therefore, CVOMT1 is likely to have evolved from EOMT1. Moreover, two genes encoding IEMT and COMT from C. breweri share 83% identity at the amino acid level, and their encoded proteins can be functionally interchangeable by mutually replacing seven amino acids (Wang and Pichersky 1998). A phylogenetic analysis has further indicated that C. breweri IEMT must have evolved recently from COMT. On the basis of an evolutionary study, Gang (2005) has reported that different genes encoding OMTs in R. hybrida have probably originated from duplication of a gene encoding COMT.
Duplicated genes are often found as tandem repeats in genomes (Ober 2005). Recently, several gene clusters involved in secondary metabolism have been identified and characterized. In Lotus japonicus, a cluster of four genes encoding chalcone isomerases has been identified on the short arm of chromosome V (Shimada et al. 2003). A three-gene cluster encoding terpene synthase on chromosome 3 of Arabidopsis has been shown to contain two genes that are identical in their coding and promoter sequences (Chen et al. 2004). However, no gene clusters encoding OMTs have been reported so far although OMTs are believed to have evolved via gene duplication followed by divergence. In addition, although cDNAs encoding OMTs have been isolated from various plants, there are few reports on genomic cloning of OMTs. To facilitate studies investigating regulation mechanism and evolution of genes encoding OMTs, it is necessary to isolate genomic clones of OMTs from different plants.
The domestic apple, Malus × domestica Borkh., belongs to the Rosaceae family. The analysis of expressed sequence tags (ESTs) from the apple has recently revealed that many genes involved in secondary metabolism are present in the apple genome (Newcomb et al. 2006). These presumably evolved by segmental duplication events and therefore are likely to be clustered (Newcomb et al. 2006). In our recent work on developing a genomewide physical map for the apple genome, we have come across some gene clusters encoding flavanone 3-hydroxylase, anthocyanidin reductase, and COMT. To date, no gene encoding COMT has been isolated from the apple. Here, we report on identifying gene clusters encoding COMT in the apple and provide evidence to support the hypothesis that plant OMTs have evolved via gene duplication followed by divergence. The evolution and duplication mechanism of genes encoding COMT in the apple are also discussed.
MATERIALS AND METHODS
Plant material and apple BAC library:
Genomic DNA, cDNA, and BAC library were all derived from the apple cultivar GoldRush. The apple BAC library was constructed using BamHI, and representing ∼5× haploid apple genome equivalents (Xu et al. 2002).
Cloning of genomic and cDNA sequences encoding COMTs in the apple:
An expressed sequence tag (EST) database from the apple was previously constructed in our lab (http://titan.biotec.uiuc.edu/cgi-bin/ESTWebsite/estima_blastui?seqSet=apple). A BLASTN search of our apple EST database for potential OMTs revealed an EST contig (ID: Apple_0223.2449.C1.Contig4283, see also Figure 1) very similar to COMT sequences from R. chinensis and Prunus dulcis; however, no sequences similar to other OMTs such as EOMT1 and CVOMT1 from O. basilicum and OOMT1 and OOMT2 from R. hybrida were identified. Several primers were designed on the basis of the contig sequence. These primers were first used to amplify genomic DNA. The authenticity of generated PCR products was first assessed by gel electrophoresis and then by sequence analysis. After a series of tests, a pair of primers, pCF (5′-GGCTGACCACTCTACCATTACC-3′) and pCR (5′-TCGAACTCCTTCTCCGTCCTC-3′) was successfully generated and used to screen the apple BAC library.
Figure 1.—
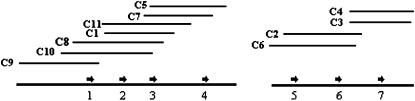
Two clusters of apple genes encoding COMT are derived from BAC clones. Arrows represent COMT genes, and are numbered 1–7. C1–C11 correspond to all positive apple BAC clones analyzed.
BAC library screening was carried out according to a PCR-based screening protocol previously described by Xu et al. (2002). The PCR program consisted of 34 cycles of 30 sec at 94°, 40 sec at 65°, 1 min at 72°, and a final extension for 10 min at 72°. Positive BAC clones were then selected for subcloning. BAC DNA was extracted from a 300-ml culture using the Plasmid Midi kit (QIAGEN, Valencia, CA). Purified BAC DNA was partially digested with the restriction enzyme Sau3Al. Digested fragments of ∼10 kb in size were harvested from a 1% agarose gel using a QIAEX II gel extraction kit (QIAGEN), and then ligated into a BamHI-digested pBlueScript-SK(+) vector. Transformation was conducted by electroporation using a Bio-Rad (Hercules, CA) gene pulser. The sequencing of positive subclones was performed according to the primer-walking method.
To obtain cDNA sequences missing from 5′ ends of genes, rapid amplification of cDNA ends (RACE) was carried out using the BD SMART RACE cDNA Amplification Kit (BD Biosciences). A gene-specific primer (5′-TCAAGCAATGC TCGTCACTCCAGTC-3′) was designed on the basis of conserved sequences of the last exon. cDNA amplification was conducted according to the manufacturer's instructions using apple flower RNA as a template. Amplification products were separated on 1% agarose. cDNA fragments were excised from the gel, purified using a QIAEX II gel extraction kit (QIAGEN), and then cloned into a pCRII-TOPO vector using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA).
Investigating physical map relationships of multiple copies of COMT genes in the apple:
Physical map relationships among the positive BAC clones were first identified using the following BAC fingerprinting method. BAC clones were grown overnight at 37° in a 3.0-ml LB medium containing chloramphenicol (12.5 μl/ml). The BAC DNA was extracted using a mini-alkaline lysis procedure. Each DNA sample was digested with 2 units BamHI for 3 hr at 37°. Restriction fragments were separated by agarose gel electrophoresis on custom Latitude HT 121-well precast gels (Cambrex) at 60 V for 15.5 hr with buffer recirculation at 4°. DNA fragments were stained using SYBR Green I (Invitrogen), and visualized by fluorescence using a Typhoon 8600 Imager (Amersham Biosciences, Piscataway, NJ). Captured gel images were analyzed using IMAGE V3.10b (http://www.sanger.ac.uk/Software/Image/). Fingerprint data were used to assemble contigs using the program FPC v7.2 (Nelson et al. 2005, http://www.agcol.arizona.edu/software/fpc). To further verify the reliability of contigs and further merge contigs, PCR-based probes derived from BAC end sequences were tested against all clones within these contigs.
Southern blot analysis for genomic DNA and BAC DNA:
Genomic DNA was extracted from young apple (cv. GoldRush) leaves using a CTAB-based protocol (Dellaporta et al. 1983). BAC DNA was extracted as described above. Five micrograms genomic DNA and ∼20 ng BAC DNA per clone were digested with BamHI and separated on 0.8% agarose gels. After treating the gel with 0.25 m HCl for 10 min and rinsing it with dd H2O, DNA was transferred to positively charged nylon membranes (Hybond-N, Amersham, UK) using the capillary transfer method. The membrane containing the transferred DNA was briefly soaked in a neutralization solution (0.2 m Tris-Cl, pH 7.5, 2× SSC) and was crosslinked by 3 min of UV irradiation on a transilluminator (312 nm). DNA blots were prehybridized with the DIG Easy Hyb (Roche, Indianapolis).
DNA probes were prepared using the PCR DIG Probe Synthesis Kit (Roche) according to the manufacturer's instructions. The primers were the same as those used above for BAC library screening. Template DNA was isolated from cDNA clones of Mdomt1. The probe used was 467 bp in size and corresponding to the 3′ region of the cDNA sequence (Figure 1). The labeled probe was added to the prehybridization solution and incubated at 42° for 16 hr. Blots were washed once with a low-stringency buffer (2× SSC containing 0.1% SDS) for 10 min at room temperature and twice with a high-stringency buffer (0.5× SSC containing 0.1% SDS) for 15 min at 65°. Then, blots were exposed to a Lumi-Film X-ray film (Hyperfilm, Amersham) at room temperature for 20 min.
RT–PCR analysis:
Genes encoding COMT in the apple genome were given the designation Mdomt. cDNA sequences of Mdomt1 and Mdomt3 were highly similar to each other. Moreover, their transcription termination sites were different from three other Mdomt's, including Mdomt2, Mdomt4, and Mdomt5. On the basis of this latter difference, a pair of primers (forward, 5′-CGACTGGAGTGACGAGCATTG-3′; reverse, 5′-CACACAACAATACAAAGAGTCATAATTC-3′) was designed to study the collective expression of the pair Mdomt1/Mdomt3. Likewise, cDNA sequences of Mdomt2 and Mdomt4 were nearly identical, and their collective expression was investigated. The primers (forward, 5′-CTCCCTCTCTCCCCCCACCA-3′; reverse, 5′-CAGAGAGAACCAATGGAAGCAC-3′) were designed on the basis of the difference in the 5′-UTR between the two pairs Mdomt2/Mdomt4 and Mdomt1/Mdomt3. In addition, a 12-bp deletion in exon 2 of Mdomt5 was observed. Forward (5′-GATTCTTCCACTGCCACAGG-3′) and reverse (5′-CACTCCAGCAACTTACGTTCC-3′) primers covering the deletion site were designed for studying the expression of Mdomt5. RT–PCR analysis was performed using a two-step procedure as previously described (Han et al. 2007). The PCR program consisted of 34 cycles (30 sec at 94°, 30 sec at 60°, 1 min at 72°, and a final extension at 72° for 10 min).
Phylogenetic analyses:
Nucleotide sequences of coding regions were aligned using CLUSTAL X (Jeanmougin et al. 1998) and adjusted manually, as necessary. The resulting data matrix was analyzed using equally weighted maximum parsimony (MP). MP trees were sought using the heuristic search strategies of PAUP* v. 4.0 (Swofford 2003). Heuristic MP searches were replicated 1000 times with random stepwise addition of taxa, tree-bisection-reconnection (TBR) branch swapping, and saving multiple trees (MulTrees). Bootstrap values (Felsenstein 1985) were calculated from 100 replicate analyses using TBR branch swapping and 500 times with stepwise addition of taxa. Only those values compatible with the 50% majority-rule consensus tree were recorded.
RESULTS
Gene clusters encoding COMT in the apple:
A total of 11 positive BAC clones, designated C1–C11, were identified from the BAC library. These BAC clones were subjected to fingerprinting analysis, and three small BAC contigs were assembled from the fingerprinting data. The first contig was ∼280 kb in size, consisting of seven clones, C1, C5, C7, C8, C9, C10, and C11 (Figure 1). The second contig consisted of two BAC clones, C2 and C6. The remaining two clones, C3 and C4, belonged to the third contig. To verify the reliability of these contigs, both ends of BAC clones, C1, C5, C9, and C3 were sequenced using T7 and SP6 primers. Of the eight BAC end sequences, one T7 end sequence from the C5 clone was highly similar to cDNA sequences of COMTs from both R. chinensis and P. dulcis, suggesting that one copy of an apple COMT encoding gene was located at this BAC end. Thus, a total of seven PCR probes were designed on the basis of these BAC end sequences and were tested against all the 11 BAC clones. The results not only clearly indicated that the contigs were correctly assembled, but also revealed a potential for merging of the second and third contigs (Table 1). As a result, two clusters encoding COMT were identified in the apple genome (Figure 1).
TABLE 1.
Presence/absence of PCR products of BAC clones related to COMT using PCR probes derived from BAC end sequences
| BAC clone
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR probe | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | C11 |
| C1-T7 | + | − | − | − | − | − | − | + | − | + | + |
| C1-SP6 | + | − | − | − | + | − | + | − | − | − | + |
| C3-SP6 | − | − | + | + | − | − | − | − | − | − | − |
| C3-T7 | − | + | + | + | + | − | − | − | − | − | |
| C5-SP6 | − | − | − | − | + | − | − | − | − | − | − |
| C9-SP6 | − | − | − | − | − | − | − | − | + | − | − |
| C9-T7 | − | − | − | − | − | − | − | + | + | + | − |
The PCR probe suffixed with T7 or SP6 indicted that the probe was designed on the basis of T7 end sequence and SP6 end sequence, respectively. +, presence of PCR products; −, absence of PCR products. C1–C11 correspond to all positive apple BAC clones analyzed.
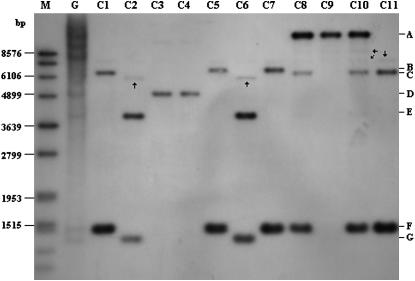
Southern blot analysis showed that all BAC clones yielded seven different bands, designated A–G (Figure 2). PCR products amplified from genomic DNA of cv. GoldRush and all positive BAC clones using the same primers as described for probe preparation cannot be digested with BamHI (data not shown). Meanwhile, none of the five apple COMT genes, Mdomt1–Mdomt5, contained the recognition site for BamHI in the region covering the probe sequence (Figure 3). These results demonstrated that a single band on the Southern blot likely corresponded to a gene copy, thus suggesting these BAC clones contained seven copies of genes encoding COMT. In one cluster, consisting of BAC clones C1, C5, C7, C8, C9, C10, and C11, there were four copies of COMT; while, in another cluster, consisting of BAC clones C2, C3, C4, and C6, there were three copies of COMT (Figure 1). The gene copy number of each BAC clone ranged from 1 to 3 (Figure 2). Moreover, these Southern blot results were consistent with the order of BAC clones in each of the two clusters. For example, each of the overlapping BAC clones C8, C9, and C10 (Figure 1) contained an “A” band (Figure 2); while, each of the overlapping BAC clones C1, C5, C7, C8, C10, and C11 (Figure 1) contained an “F” band (Figure 2). As a result, blotted bands of BAC clones, A, B C, D, and F, putatively corresponded to COMT genes 1, 4, 2, 7, and 3 noted in Figure 1; while blot bands E and G corresponded to COMT genes 5 and 6 noted in Figure 1. In addition, the apple genomic DNA exhibited three additional bands to those identified in positive BAC clones (Figure 2), thus suggesting that there might be more than seven copies of genes encoding COMT in apple.
Figure 2.—
Southern blot analysis of genomic DNA isolated from apple leaf tissues. M, standard DNA marker; G, apple genomic DNA; and C1–C11 correspond to 11 positive BAC clones. The faint bands are indicated by arrows. Letters on the left indicate hybridizing bands of different sizes. A, B C, D, and F bands of BACs correspond to nos. 1, 4, 2, 7, and 3 of COMT genes in Figure 1. E or G bands of BACs correspond to no. 5 or 6 of COMT genes in Figure 1. F band of C8, D band of C3, F band of C5, E band of C2, and G band of C2 correspond to Mdomt1, Mdomt2, Mdomt3, Mdomt4, and Mdomt5, respectively.
Figure 3.—
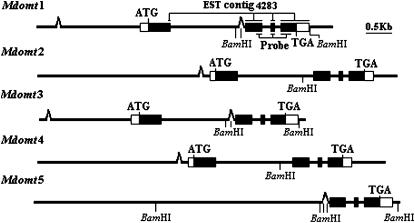
Structure of the apple Mdomt genes. Solid boxes indicate exons and open boxes represent 5′- and 3′-UTRs. The transposon-like elements are indicated by open triangles.
Classification of genes encoding COMT in the apple:
Sequence alignments of clones of PCR products amplified from cv. GoldRush using primers pCF/pCR revealed two single-nucleotide polymorphisms (SNPs), including a T/A SNP in intron 2 and a T/G SNP in exon 4. The two SNPs disrupted the TaqI recognition site, TCGA, to either ACGA or GCGA. Thus, PCR products amplified from all 11 positive BAC clones using primers pCF/pCR were subjected to digestion with TaqI and then separated on a 1% agarose gel. Three different sizes of DNA bands were observed and were designated 3T, 3M, and 3B corresponding to top (596 bp), middle (510 bp), and bottom (453 bp) bands, respectively (data not shown). The numerical 3′ indicated that the cleaved amplified polymorphic sequence (CAPS) marker was developed from the 3′ region of genes encoding COMT. Seven BAC clones, C1, C5, C7, C8, C9, C10, and C11, generated a single 3B band. Two BAC clones, C3 and C4, generated a single 3M band, while the remaining two BAC clones, C2 and C6, generated all three bands, 3T, 3M and 3B. The middle-sized band, 3M, of C2 and C6 was later proved to be a PCR artifact using the following methods. First, Southern blotting indicated that both C2 and C5 contained two copies of COMT genes as they exhibited two strong bands (Figure 2). Second, no subclone of C2 corresponding to the middle-sized band was identified. Third, when PCR products generated from the two subclones of C2, corresponding to high and low bands, respectively, were mixed, denatured, and annealed, using the same PCR program previously used to identify positive BAC clones, and digested with TaqI, following electrophoresis, three bands, including the middle-sized band, were detected (data not shown).
Meanwhile, 5′-cDNA sequences of apple genes encoding COMT were recovered using 5′-end RACE analysis, and a (CT)n repeat in the 5′-UTR region was found. A pair of primers flanking the repeat sequence (forward, 5′-CCACTTAGCACACACCATCTC-3′; reverse, 5′-CAGAGAGAACCAATGGAAGCAC-3′) was then designed. The primers were used to amplify all positive BAC clones, and three different sizes of bands were displayed (data not shown). Since the repeat sequence was located in the 5′-UTR, the top-, middle-, and bottom-sized bands were designated as 5T, 5M, and 5B, respectively. Therefore, results of both microsatellite polymorphism and CAPS marker analysis indicated that there were five types of Comt genes in the apple genome. BAC clones C1, C8, C9, C10, and C11 contained type I (5M3B); C3 and C4 comprised type II (5B3M); C5 and C7 had type III (5T3B); while, C2 and C6 consisted of types IV (5B3T) and V (5B3B), respectively.
Genomic structure and sequence characteristics of the apple genes encoding COMT:
To isolate the five identified types of genes encoding COMT, BAC clones C2, C3, C5, and C8 were selected and subcloned. Full genomic DNA sequences for four genes encoding COMT were successfully recovered. They were designated as Mdomt1, Mdomt2, Mdomt3, and Mdomt4, corresponding to types I, II, III, and IV, respectively. Alignment of genomic DNA sequences with cDNA sequences revealed that apple Mdomt genes consisted of four exons and three introns (Figure 3). Border sequences of all three introns conformed to the GT-AG rule. All four copies of Mdomt genes contained a microsatellite repeat at the 5′-UTR located immediately upstream of the start codon (Figure 4). The coding sequences of Mdomt1, Mdomt2, Mdomt3, and Mdomt4 were all 1095 bp in size, and the putative protein consisted of 365-amino-acid residues.
Figure 4.—
Repeat sequences at the 5′-UTR of apple Mdomt genes. The start codons are shaded. Dashes indicate sequence gap. Sequences different from those of the top line are indicated in lowercase letters.
A genomic DNA sequence of ∼7.2 kb in size corresponding to a type V Mdomt gene was also isolated from the end of BAC clone C2 and designated as Mdomt5. When the genomic sequence of Mdomt5 was aligned with cDNA sequences of other copies of Mdomt genes, it covered exons 2–4, but did not include exon 1. A long fragment of ∼6 kb in size, from the beginning of the genomic DNA sequence to the putative exon 2, had no match to Mdomt cDNA sequences. However, as a part of this fragment was highly similar to intron 1 of other Mdomt genes, this was designated as intron 1 in Mdomt5.
Interestingly, comparisons of genomic DNA sequences of the five Mdomt genes revealed two transposable element-like sequences. The first transposon-like sequence was located in the promoter region and was present in all four Mdomt genes, Mdomt1–Mdomt4 (Figure 5). The transposon-like sequence among the four genes showed >85% identity and contained a 21-nucleotide terminal inverted repeat sequence, 5′-GGGGTGTGATATCCACACACC—GGTGTGTGGATAGCACACCCC-3′. Target site sequences of the transposon in the two pairs Mdomt1/Mdomt3 and Mdomt2/Mdomt4 were TAATACAA and TAAATAAAA, respectively (Figure 5). The second transposon-like sequence was inserted in intron 1 of Mdomt genes and was very close, by 49 nucleotides, to exon 2 (Figure 6). This second transposon-like sequence was clearly present in Mdomt5. It also generated a 9-nucleotide direct repeat sequence, TTGTAGATT, at the target insertion site, and contained a 10-nucleotide inverted repeat sequence 5′-GGAAATGGAT—ATCCCTTTCC-3′ (Figure 6). This second transposon-like sequence was also identified in both Mdomt1 and Mdomt3 although this sequence showed slight variations between the pair Mdomt1/Mdomt3 and Mdomt5 (Figure 6). However, this sequence was not detected in either Mdomt2 or Mdomt4 although the transposon-target insertion site TTGTAGATT was found in these two genes. These results strongly demonstrated that the second transposon-like sequence was actually a transposable element although it was not clear if the transposon was present earlier in Mdomt2 and Mdomt4, but was excised at some later time leaving the footprint sequence TTGTAGATT behind.
Figure 5.—
Transposon-like sequences in the promoter region of Mdomt genes. Letters in boldface type indicate repeated sequences of the insertion site. Palindrome sequences are underlined. The dashes indicate sequence gaps. Sequences different from those of the top line are indicated in lowercase letters.
Figure 6.—
Transposon-like sequences in intron 1 of Mdomt genes. Insertion and/or excision site sequences are indicated in boldface type. The start sequence of exon 2 is shadowed. Dashes indicate sequence gap. Sequences different from those of the top line are indicated in lowercase letters. The inverted sequences are underlined.
Expression of Mdomt genes:
Expression of all Mdomt genes was investigated by RT–PCR analysis although the complete cDNA sequence of Mdomt5 was not available. As cDNA sequences of Mdomt1 and Mdomt3 showed 98% identity, it was difficult to individually study their expression profiles. Therefore, their collective expression was investigated. Similarly, cDNA sequences of Mdomt2 and Mdomt4 showed 99% identity, and therefore, their collective expression was analyzed as well.
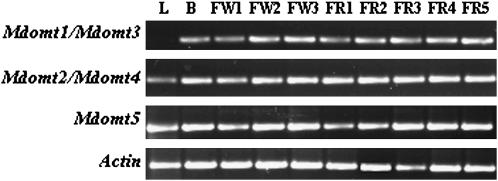
The collective transcripts of Mdomt1/Mdomt3 were detected in buds, flowers, and fruits, but not in leaves, while the collective transcripts of Mdomt2/Mdomt4 accumulated in all tissues, including leaves, buds, flowers, and fruits (Figure 7). Moreover, Mdomt5 was expressed in all tissues analyzed (Figure 7).
Figure 7.—
Analysis of expression profiles of the Mdomt genes by RT–PCR. Lanes correspond to the following: L, leaves; B, buds; FW1, flower buds at the pink stage (first pink); FW2, flower buds at the balloon stage (full pink); FW3, flowers at full bloom (open flowers); FR1, young fruitlet I (9 days after pollination [DAP]; FR2, young fruitlet II (16 DAP); FR3, young fruitlet III (44 DAP); FR4, maturing fruit I (104 DAP); FR5, ripe fruit (166 DAP). (Bottom) Loading control of actin transcripts in each sample by RT−PCR.
Evolution of Mdomt genes in the apple genome:
Mdomt2, Mdomt4, and Mdomt5 were isolated from BAC clones C3, C2, and C2, respectively, and they were clustered as described above (Figure 1). Southern blots of genomic DNA indicated that the hybridizing fragment of C3 was different in size from those of C2 (Figure 2). Meanwhile, sequences from 5′- and 3′-flanking regions of Mdomt2 and Mdomt4 had no significant matches. These results suggested that Mdomt2 and Mdomt4 were indeed two independent genes. However, sequence alignment of Mdomt2 and Mdomt4 revealed presence of a nearly identical region ranging from 2761 bp upstream of the putative start codon to 219 bp downstream of the putative end codon. This region showed 98% nucleotide sequence identity between the two clustered genes Mdomt2 and Mdomt4, suggesting these were derived from a recent duplication event. Sequence comparisons of Mdomt5 and the nearly identical pair Mdomt2/Mdomt4 also showed two common homologous regions (>96% identity). The first homologous region spanned from the transposon target site in intron 1 of Mdomt2/Mdomt4 to 219 bp downstream of the putative end codon. It was apparent that a sequence divergence point at the 3′ end of Mdomt2, Mdomt4, and Mdomt5 was located 219 bp downstream of the putative end codon. The second homologous region was identified in intron 1. A small fragment of 143 bp in size upstream of the transposable element-like sequence in intron 1 of Mdomt5 almost matched exactly against sequences upstream of the target insertion site TTGTAGATT in intron 1 of both Mdomt2 and Mdomt4 (Figure 6). The two homologous regions identified in Mdomt2, Mdomt4, and Mdomt5 suggested that these three clustered genes were derived from a common ancestor.
Mdomt1 and Mdomt3 were derived from the two overlapped BAC clones C8 and C5, respectively (Figure 1). Complete DNA sequences of Mdomt1 and Mdomt3 showed 97% identity. Sequence variations were mainly caused by short stretches of duplication/deletion that were observed in both 5′-UTR and intron 1. When the two genomic clones of Mdomt1 and Mdomt3 were digested in silico with BamHI, they yielded an equal fragment of 1.4 kb in size at the 3′ end. Genomic DNA and BAC DNA were digested with BamHI for Southern blot analysis and were hybridized with probes corresponding to the 3′-cDNA end. Genomic DNA and six overlapping BAC clones, including C1, C5, C7, C8, C10, and C11 showed the same 1.4-kb band (Figures 1 and 2). These results, along with the fact that the apple is heterozygous due to self-incompatibility, indicated that Mdomt1 and Mdomt3 were allelic. When DNA sequences of these two alleles were further compared with those of the three other Mdomt genes, several striking similar features were observed. First, coding region sequences of all five Mdomt genes shared >97% identity. Second, introns 2 and 3 among the five Mdomt genes shared >96 and 97% identities, respectively. The first intron showed ∼71% sequence identity between the two pairs of Mdomt1/Mdomt3 and Mdomt2/Mdomt4. Third, intron 1 of Mdomt5 was larger than that found in the remaining four Mdomt genes, but a small fragment of 143 bp in size upstream of the transposable element-like sequence was nearly equal in size to those found in all other four Mdomt genes (Figure 6). Finally, the transposon-like sequence in intron 1 of Mdomt5 was also present at the same location as that found in both Mdomt1 and Mdomt3 in spite of a deleted sequence observed in both Mdomt1 and Mdomt3 (Figure 6). These striking similar features unambiguously suggested that the five genes, Mdomt1–Mdomt 5, were derived from a common ancestor.
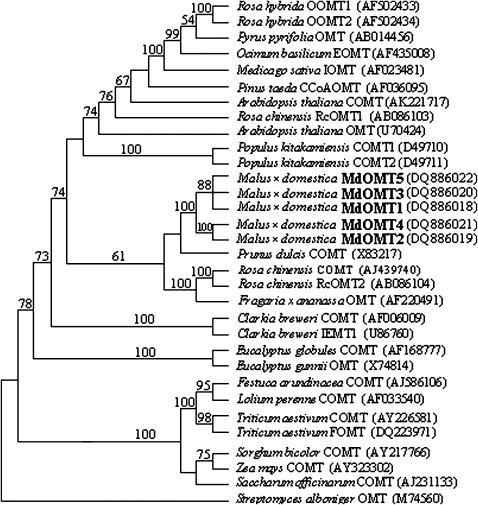
Phylogenetic analysis using the maximum-parsimony method revealed that all five sequences encoding COMT in the apple are most closely related to those from other Rosaceae species such as P. dulcis, R. chinensis, and Fragaria × ananassa (Figure 8). This supported the fact that Mdomt1–Mdomt 5 in the apple are clearly members of the COMT family. Sequences of Mdomt1, Mdomt3, and Mdomt5 are grouped together as a sister clade to the other two sequences of Mdomt2 and Mdomt4, further confirming that all five Mdomt genes in the apple were derived from a common ancestor. Moreover, the two transponson-like sequences in Mdomt genes were blasted against NCBI database (http://www.ncbi.nlm.nih.gov/), and significant hits were detected only in the apple genes. In addition, microsatellite repeats in the 5′-UTR of Mdomt genes were not detected in any genes encoding COMT from other plant species. The two transposon-like sequences and microsatellite sequences were uniquely found in the apple Mdomt genes, suggesting the duplication of Mdomt genes likely occurred during and after the speciation of apple.
Figure 8.—
Phylogenetic tree of five apple COMTs and other species OMTs. Numbers on branches represent bootstrap values; those values <50% are not indicated. OMT data were obtained from GenBank with accession numbers given in parentheses. The apple COMTs are indicated in boldface type.
DISCUSSION
cDNA sequences of genes encoding COMT have been isolated from a variety of species, and COMTs were previously reported to be encoded by a small gene family in plants. For example, Hayakawa et al. (1996) reported that the copy number of genes encoding COMT in Populus kitakamiensis was less than three; while two copies of COMT encoding genes were isolated in Zea mays (Guillet-Claude et al. 2004). However, two clusters, consisting of seven copies of genes encoding COMT, were identified in the present study. In addition to the multiple copies of COMT, two alleles, Mdomt1 and Mdomt3, were also identified in apple. The presence of multiple copies and alleles suggested that genes encoding COMT in the apple genome did not belong to a small gene family. This conclusion was consistent with recent findings, based on analysis of apple ESTs, that high copy numbers of genes involved in secondary metabolism were frequent in the apple genome (Newcomb et al. 2006). Moreover, our Southern blot analysis clearly indicated that there were more than seven copies of genes encoding COMT in the apple. Failure in capturing all members of the COMT gene family in apple might be attributed to genome coverage of the apple BAC library, representing ∼5× haploid apple genome equivalents, used for screening of positive BAC clones. On the other hand, positive BAC clones showed presence of several faint bands likely corresponding to additional genomic DNA bands (Figure 2). However, the possibility that these additional bands might be nonspecific bands cannot be completely ruled out.
In this study, five clones encoding COMT in the apple have been isolated. Of these five clones, four contain full-coding sequences. The four genes, Mdomt1–Mdomt4, show significant sequence identities. It is well known that only minor changes in sequences of genes encoding COMT are often sufficient to completely alter their substrate and product specificities (Wang and Pichersky 1998; Gang et al. 2002). Thus, sequence identity is insufficient to predict the function of each of these newly identified genes coding for COMT, and further studies are needed to determine the functions of these various COMT genes in the apple genome.
Genes encoding COMT in the apple are expanded by segmental duplication:
Gene duplication is assumed to be a major driving force for recruitment of genes for secondary metabolism (Pichersky and Gang 2000). Polyploidy (genome duplication) is a significant evolutionary process in higher organisms, and genomes of flowering plants are reported to have incurred one or more polyploidization events during evolution (Masterson 1994). Therefore, gene duplication in plants may arise from polyploidization (genome duplication) and/or segmental duplication. In this study, two clusters encoding COMT have been identified in the apple genome. One cluster contains three copies, and all have been isolated; while another cluster consists of four copies, and two alleles have been isolated. The analysis of genomic DNA sequences revealed that the five genes encoding COMT in the apple are distinguished by a (CT)n microsatellite in the 5′-UTR and two transposon-like sequences in the promoter region and intron 1. Discovery of the transposon-like sequence in intron 1 unambiguously traced the five Mdomt genes back to a common ancestor. Moreover, introns also showed several similar features among the five Mdomt genes. First, both introns 2 and 3 showed >96% nucleotide sequence identities among the five genes. Second, nucleotide sequences of intron 1 showed ∼70% identity between the two pairs Mdomt1/Mdomt3 and Mdomt2/Mdomt4. Finally, a small fragment upstream of the transposon in intron 1 of Mdomt5 was almost identical to that of the two pairs Mdomt1/Mdomt3 and Mdomt2/Mdomt4. These similarities in intron sequences further supported the hypothesis that duplication of Mdomt genes must have occurred more recently and within a relatively short time period during evolution of the apple.
Although only a single pair of alleles was isolated from one cluster, consisting of four copies of genes encoding COMT, analysis of a CAPS marker used to classify positive BAC clones showed that all four copies within this cluster were of the same genotype (Figure 3). This result coupled with the fact that these four copies were clustered suggested that all four copies were also generated from a common progenitor.
All the above results suggest that an initial duplication of an Mdomt ancestor has resulted in the generation of two progenitors of the two clusters, although further studies are needed to address the physical relationship between the two clusters encoding COMT. The two progenitors must have subsequently undergone further duplication, resulting in the two clusters identified in this study.
The two transposon-like sequences found in Mdomt genes in this study have not been identified in previously reported genes encoding COMT in other closely related tree species such as Populus and Prunus or any other plants. In addition, the microsatellite sequence found in the 5′-UTR of Mdomt genes has also not been found in previously reported COMT cDNA sequences in plants. Therefore, these results along with the finding that clusters are derived from a common ancestor all suggest that multiple copies of COMT in the apple genome are likely derived from segmental duplication during and after speciation of the apple rather than from polyploidization of the apple. In Arabidopsis, gene copies encoding regulatory proteins such as transcription factors and signal transduction proteins tend to retain when generated by polyploidization; whereas these genes are rapidly lost when derived from segmental duplication (Maere et al. 2005). By contrast, gene copies involved in secondary metabolism or abiotic stress are likely to be retained when derived from segmental duplication; whereas duplicated gene copies following whole-genome duplications are more rapidly lost (Maere et al. 2005). Thus, the pattern of evolution in Arabidopsis seems to also be conserved in the apple. In addition, the discovery of the evolutionary pathway of apple Mdomt genes provides, for the first time, direct molecular evidence supporting previous speculations that functionally distinct OMTs may have evolved from a common ancestral gene through gene duplication and subsequent divergence (Gang 2005).
Possible mechanism of gene amplification:
To investigate the possible mechanism responsible for duplication of apple Mdomt genes, we focused on the 5′-flanking sequences of the two recently duplicated and clustered genes Mdomt2 and Mdomt4. The 5′-flanking regions, of 3.7 and 3.3 kb in size upstream of the putative start codon of Mdomt2 and Mdomt4, respectively, were sequenced. Sequence comparisons revealed that the sequence divergence point between Mdomt2 and Mdomt4 was located 2655 bp upstream of the putative start codon. A DNA fragment 675 bp in size, downstream of the divergence point from Mdomt2 or Mdomt4, shared >90% nucleotide sequence identity with a dispersed repeat sequence previously reported in the apple. The repeat sequence was deposited in the NCBI database with the accession no. AM167520. Therefore, the duplication of apple Mdomt genes might have resulted from unequal recombination due to the presence of homologous sequences flanking these genes.
DNA secondary structures and sequence variations in the promoter region of Mdomt genes:
Overall, the genomic DNA sequence, except for the promoter region, shows >80% identity between the two pairs Amot1/Mdomt3 and Amot2/Mdomt4. However, alignment of the promoter region sequences of the two pairs Amot1/Mdomt3 and Amot2/Mdomt4 has revealed a small homologous region covering the transposon-like sequence. Meanwhile, location of the transposon-like sequence was not comparable between the two pairs Amot1/Mdomt3 and Amot2/Mdomt4. The transposon-like sequences in the promoter region of the two pairs Amot1/Mdomt3 and Amot2/Mdomt4 were inserted 1639 and 297 bp, respectively, upstream of the putative start codon. Although the two pairs are derived from a common ancestor, as documented above, significant sequence variations must have occurred in the promoter region since the early generation of the two pairs Amot1/Mdomt3 and Amot2/Mdomt4. It has been reported that slippage caused by strand mispairing often occurred during DNA replication, and DNA regions capable of assuming hairpin-like secondary structures are particularly prone to this error (Levinson and Gutman 1987). Thus, hairpin formation may play a destabilizing role in eukaryotic genomes (Sinden 2001). Interestingly, we have identified that distal regions of the transposon-like sequence in the promoter region of Mdomt genes are capable of forming palindromic, hairpin-like structures. For example, the sequence AAAAGGGGTGTGATATCCACACACCCCTTTT at the left end of the transposon-like sequence in Amot2 and Mdomt4 possesses a 12-bp terminal inverted repeat (Figure 7). Two sequences, GGGGTGTGATATCCACACACCCC and GGGGTGTGTGGATAGCACACCCC at left and right ends, respectively, of the transposon-like sequence in both Mdomt1 and Mdomt3 consist of an 8-bp terminal inverted repeat (Figure 5). Thus, it is reasonable to speculate that sequence variations in the promoter regions of the two pairs Amot1/Mdomt3 and Amot2/Mdomt4 are mainly due to replication slippage, which is enhanced and facilitated by the hairpin-like structures present in the promoter region.
Previously, two genes encoding COMT, homt1 and homt2, were isolated from P. kitakamiensis (Hayakawa et al. 1996). Transcripts of homt1 did not accumulate in young leaves; while homt2 transcripts accumulated in all tissues analyzed (Hayakawa et al. 1996). These observed differences in gene expression were also observed in the apple Amot genes in the present study. RT–PCR analysis showed that Amot1 and Mdomt3 are not expressed in young leaves; whereas Amot2 and/or Mdomt4 are expressed in all different tissues analyzed. As the promoter region is known to play an important role in gene expression, the influence of promoter sequences on expression of genes encoding COMT has been reported in tobacco (Toquin et al. 2003). Therefore, differences in expression profiles of genes encoding COMT in the apple might be due to sequence variations in the promoter region of the two pairs Amot1/Mdomt3 and Amot2/Mdomt4. It is worthwhile to note that a (CT)n microsatellite was found in the 5′- UTR of Amot genes. Whether this repeat is actually involved in Amot gene expression must be further investigated. However, two genes encoding COMT in P. kitakamiensis exhibited different expression profiles although no repeat sequence was identified in the 5′-UTR of these two genes. This suggested that the (CT)n repeat found in the 5′-UTR of Amot genes might have little influence on expression of these genes in the apple.
What happened to intron 1 of Mdomt5?
A DNA sequence of ∼6 kb in size upstream of the putative start site of exon 2 of Mdomt5 was isolated. Of this sequence, only a small fragment, ∼400 bp in size, covering the transposon-like sequence matched against sequences of intron 1 of the other four Mdomt genes, Mdomt1–Mdomt4 (Figure 6). The remainder of this sequence, ∼5.6 kb in size, was uniquely present in Mdomt5. This unique sequence was aligned and blasted against the NCBI database. Three short sequences (∼100 bp each) from different sites of intron 1 of Mdomt5 showed >85% identities to the apple retrotransposon dem1 (GenBank accession no. AJ291492). Overall, this unique sequence of ∼5.6 kb in size showed 60% identity to that of dem1. The dem1 retrotransposon, >9 kb in size, was identified in the intron of a gene responsible for the development of floral organs in the apple (Yao et al. 2001). Therefore, it is likely that the observed variation in intron 1 of Mdomt5 may have arisen from an insertion of a large transposon, although further investigation is warranted. Moreover, a repeat sequence, (AT)27, was found in intron 1 of Mdomt5. This repeat sequence has the potential to form a secondary structure, leading to DNA variation. However, it has been reported that (TA)n repeats are widely distributed in eukaryotic genomes, and they are not likely associated with DNA instability (Kost-Alimova et al. 2003). Thus, it is likely that the (AT)n repeat plays only a small role, if any, in destabilizing intron 1 of Mdomt5.
The transposon-like sequence in intron 1 of Mdomt5 was identified at the same location as those in Mdomt1 and Mdomt3. Moreover, the CAPS marker analysis revealed that Mdomt5 shared the same genotype with the pair Mdomt1/Mdomt3 (Figure 3). These results indicated that Mdomt5 and the pair Mdomt1/Mdomt3 were derived from a common progenitor. On the other hand, a 148-bp sequence upstream of the transposon-like sequence and its target site sequence for insertion in intron 1 of Mdomt5 were exactly matched against those of the pair Mdomt2/Mdomt4 (Figure 6). This suggested that Mdomt5 and the pair Mdomt2/Mdomt4 were also derived from a common progenitor. Taken together, these results demonstrated that Mdomt5 was an intermediary between the two pairs Mdomt1/Mdomt3 and Mdomt2/Mdomt4 during the process of evolution of genes encoding COMT in the apple. To determine which pair was generated earlier, putative amino acid sequences of the two pairs Mdomt1/Mdomt3 and Mdomt2/Mdomt4 were compared with those of COMTs from species closely related to the apple. Putative amino acid sequences of Mdomt1/Mdomt3 showed 91, 89, 86, and 84% identities to those of COMTs from R. hybrida, F. × ananassa, P. kitakamiensis, and Eucalyptus globules, respectively. Moreover, putative amino acid sequences of Mdomt2/Mdomt4 showed 87, 86, 83, and 80% identities to those genes encoding COMTs from R. hybrida, F. × ananassa, P. kitakamiensis, and E. globules, respectively. As the pair Mdomt1/Mdomt3 showed slightly higher amino acid sequence identities than the pair Mdomt2/Mdomt4 to genes encoding COMTs from related species, this suggested that Mdomt2/Mdomt4 might have undergone duplication after Mdomt1/Mdomt3. Thus, it is likely that Mdomt5 corresponded to the progenitor, whose duplication generated the parent of the pair Mdomt2/Mdomt4, and this parent in turn underwent recent duplication resulting in the generation of Mdomt2 and Mdomt4. These results indicated that the observed variation in intron 1 of Mdomt5 must have occurred since the early generation of the parent for the pair Mdomt2/Mdomt4. Moreover, the transposon-like sequence was likely excised from intron 1 of the parent for the pair Mdomt2/Mdomt4 prior to its duplication and leaving the footprint TTGTAGATT intact (Figure 6).
When the partial coding sequence of Mdomt5 was aligned with those of the other four Mdomt genes, two mutations were identified in Mdomt5. The first was a 12-bp deletion in exon 2, which did not cause a frameshift. The second was an addition of a single base pair 66 bp upstream of the original putative stop codon, resulting in a new stop codon 48 bp downstream of the insertion site and a six-amino-acid deletion at the peptide terminal. RT–PCR analysis revealed that Mdomt5 was expressed in all tissues analyzed. Therefore, for the future, it is worthwhile to investigate whether Mdomt5 represents a functionally distinct gene encoding OMT.
Acknowledgments
This project was supported by the U.S. Department of Agriculture Cooperative State Research, Education and Extension Service–National Research Initiative–Plant Genome Program grant 2005-35300-15538.
References
- Chen, F., D. Ro, J. Petri, J. Gershenzon, J. Bohlmann et al., 2004. Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8-cineole. Plant Physiol. 135: 1956–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, A. B., P. L. Gregersen, C. E. Olsen and D. B. Collinge, 1998. A flavonoid 7-O-methyltransferase is expressed in barley leaves in response to pathogen attack. Plant Mol. Biol. 36: 219–227. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S. L., J. Wood and J. B. Hicks, 1983. A plant DNA mini-preparation, Version II. Plant Mol. Biol. Rep. 1: 19–21. [Google Scholar]
- Felsenstein, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Gang, D. R., 2005. Evolution of flavors and scents. Annu. Rev. Plant Biol. 56: 301–325. [DOI] [PubMed] [Google Scholar]
- Gang, D. R., N. Lavid, C. Zubieta, F. Chen, T. Beuerle et al., 2002. Characterization of phenylpropene O-methyltransferases from sweet basil: facil change of substrate specificity and convergent evolution within a plant O-methyltrasferase family. Plant Cell 14: 505–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier, A., P. J. Gulick and R. K. Ibrahim, 1998. Characterization of two cDNA clones which encode O-methyltransferases for the methylation of both flavonoid and phenylpro panoid compounds. Arch. Biochem. Biophys. 351: 243–249. [DOI] [PubMed] [Google Scholar]
- Guillet-Claude, C., C. Birolleau-Touchard, D. Manicacci, M. Fourmann, S. Barraud et al., 2004. Genetic diversity associated with variation in silage corn digestibility for three O-methyltransferase genes involved in lignin biosynthesis. Theor. Appl. Genet. 110: 126–135. [DOI] [PubMed] [Google Scholar]
- Han, Y., K. Gasic, F. Sun, M. Xu and S. S. Korban, 2007. A gene encoding starch branching enzyme I (SBEI) in the apple (Malus × domestica, Rosaceae) and its phylogenetic relationship to Sbe genes from other angiosperms. Mol. Phylogenet. Evol. 43: 852–863. [DOI] [PubMed] [Google Scholar]
- Hayakawa, T., K. Nanto, S. Kawai, Y. Katayama and N. Morohoshi, 1996. Molecular cloning and tissue-specific expression of two genes that encode caffeic acid O-methyltransferases from Populus kitakamiensis. Plant Sci. 113: 157–165. [Google Scholar]
- He, X., J. T. Reddy and R. A. Dixon, 1998. Stress responses in alfalfa (Medicago sativa L.). XXII. cDNA cloning and characterization of an elicitor-inducible isoflavone 7-O-methyltransferase. Plant Mol. Biol. 36: 43–54. [DOI] [PubMed] [Google Scholar]
- Ibrahim, R. K., A. Bruneau and B. Bantignies, 1998. Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol. Biol. 36: 1–10. [DOI] [PubMed] [Google Scholar]
- Inoue, K., V. J. Sewalt, G. B. Murray, W. Ni, C. Sturzer et al., 1998. Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol. 117: 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins and T. J. Gibson, 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23: 403–405. [DOI] [PubMed] [Google Scholar]
- Kost-Alimova, M., H. Kiss, L. Fedorova, Y. Yang, J. P. Dumanski et al., 2003. Coincidence of synteny breakpoints with malignancy-related deletions on human chromosome 3. Proc. Natl. Acad. Sci. USA 100: 6622–6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavid, N., J. Wang, M. Shalit, I. Guterman, E. Bar et al., 2002. O-Methyltransferases involved in the biosynthesis of volatile phenolic derivatives in rose petals. Plant Physiol. 129: 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson, G., and G. A. Gutman, 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4: 203–221. [DOI] [PubMed] [Google Scholar]
- Li, L., J. Popko, X. Zhang, K. Osakabe, C. J. Tsai et al., 1997. A novel multifunctional O-methyltransferase implicated in a dual methylation pathway associated with lignin biosynthesis in loblolly pine. Proc. Natl. Acad. Sci. USA 94: 5461–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere, S., S. De Bodt, J. Raes, T. Casneuf, M. Van Montagu et al., 2005. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102: 5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson, J., 1994. Stomatal size in fossil plants: Evidence for polyploid in majority of angiosperms. Science 264: 421–424. [DOI] [PubMed] [Google Scholar]
- Maury, S., P. Geoffroy and M. Legrand, 1999. Tobacco O-methyltransferases involved in phenylpropanoid metabolism: The different caffeoyl-coenzyme A/5-hydroxyferuloyl-coenzyme A 3/5-O-methyltransferase and caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol. 121: 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, W. M., A. K. Bharti, E. Butler, F. Wei, G. Fuks et al., 2005. Whole-genome validation of high-information-content fingerprinting. Plant Physiol. 139: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb, R. D., R. N. Crowhurst, A. P. Gleave, E. H. A. Rikkerink, A. C. Allan et al., 2006. Analyses of expressed sequence tags from apple. Plant Physiol. 141:147–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober, D., 2005. Seeing double: gene duplication and diversification in plant secondary metabolism. Trends Plant Sci. 10: 444–449. [DOI] [PubMed] [Google Scholar]
- Pichersky, E., and D. R. Gang, 2000. Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci. 5: 439–445. [DOI] [PubMed] [Google Scholar]
- Schwab, W., 2003. Metabolome diversity: Too few genes, too many metabolites? Phytochemistry 62: 837–849. [DOI] [PubMed] [Google Scholar]
- Shimada, N., T. Aoki, S. Sato, Y. Nakamura, S. Tabata et al., 2003. A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol. 131: 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden, R. R., 2001. Neurodegenerative diseases. Origins of instability. Nature 411: 757–758. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA.
- Toquin, V., B. Grausem, P. Geoffroy and M. Legrand, 2003. Structure of the tobacco caffeic acid O-methyltransferase (COMT) II gene: identification of promoter sequences involved in gene inducibility by various stimuli. Plant Mol. Biol. 52: 495–509. [DOI] [PubMed] [Google Scholar]
- Wang, J., and E. Pichersky, 1998. Characterization of S-adenosyl-L-methionine: (iso)eugenol O-methyltransferase involved in floral scent production in Clarkia breweri. Arch. Biochem. Biophys. 349: 153–160. [DOI] [PubMed] [Google Scholar]
- Wang, J., N. Dudareva, S. Bhakta, R. A. Raguso and E. Pichersky, 1997. Floral scent production in Clarkia breweri (onagraceae). II. Localization and developmental modulation of the enzyme S-adenosyl-L-methionine: (iso)eugenol O-methyltransferase and phenylpropanoid emission. Plant Physiol. 114: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., N. Watanabe, S. Mita, Y. Ueda, M. Shibuya et al., 2003. Two O-Methyltransferases isolated from flower petals of Rosa chinensis var. spontanea involved in scent biosynthesis. J. Biosci. Bioeng. 96: 119–128. [PubMed] [Google Scholar]
- Xu, M., S. S. Korban, J. Song and J. Jiang, 2002. Constructing a bacterial artificial chromosome library of the apple cultivar Goldrush. Acta Hortic. 595: 103–112. [Google Scholar]
- Yao, J., Y. Dong and B. A. Morris, 2001. Parthenocarpic apple fruit production conferred by transposon insertion mutations in a MADS-box transcription factor. Proc. Natl. Acad. Sci. USA 98: 1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Z., and J. Varner, 1995. Differential expression of two O-methyltransferases in lignin biosynthesis in Zinnia elegans. Plant Physiol. 108: 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]