Figure 1. Model for PriB function based on structural similarity to SSB.
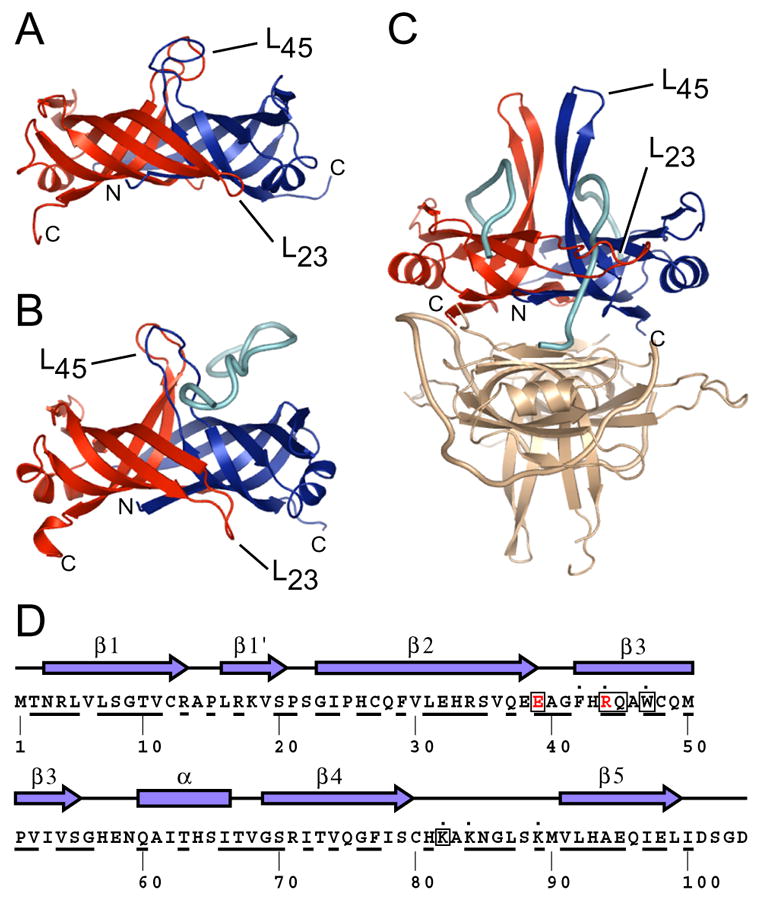
Ribbon diagrams of crystal structures of (A) apo PriB (Protein Databank accession code 1V1Q) (Liu et al., 2004), (B) PriB:ssDNA (Protein Databank accession code 2CCZ) (Huang et al., 2006), and (C) SSB:ssDNA (Protein Databank accession code 1EYG) (Raghunathan et al., 2000) were rendered with Pymol (DeLano, 2002) and are colored as follows: chain A (PriB and SSB), red; chain B (PriB and SSB), blue; ssDNA, cyan. One homodimer of the SSB:ssDNA homotetramer is colored beige. The amino- and carboxy-termini of chains A and B for PriB and SSB are indicated, as are the L23 and L45 loops. (D) The secondary structural elements of PriB are shown above the primary sequence and residues that are highly conserved among sequenced priB homologs are underlined. PriB residues are indicated that are important for interactions with PriA (red), DnaT (boxed), and ssDNA (dot).
