Figure 2. PriB physically interacts with the helicase domain of PriA.
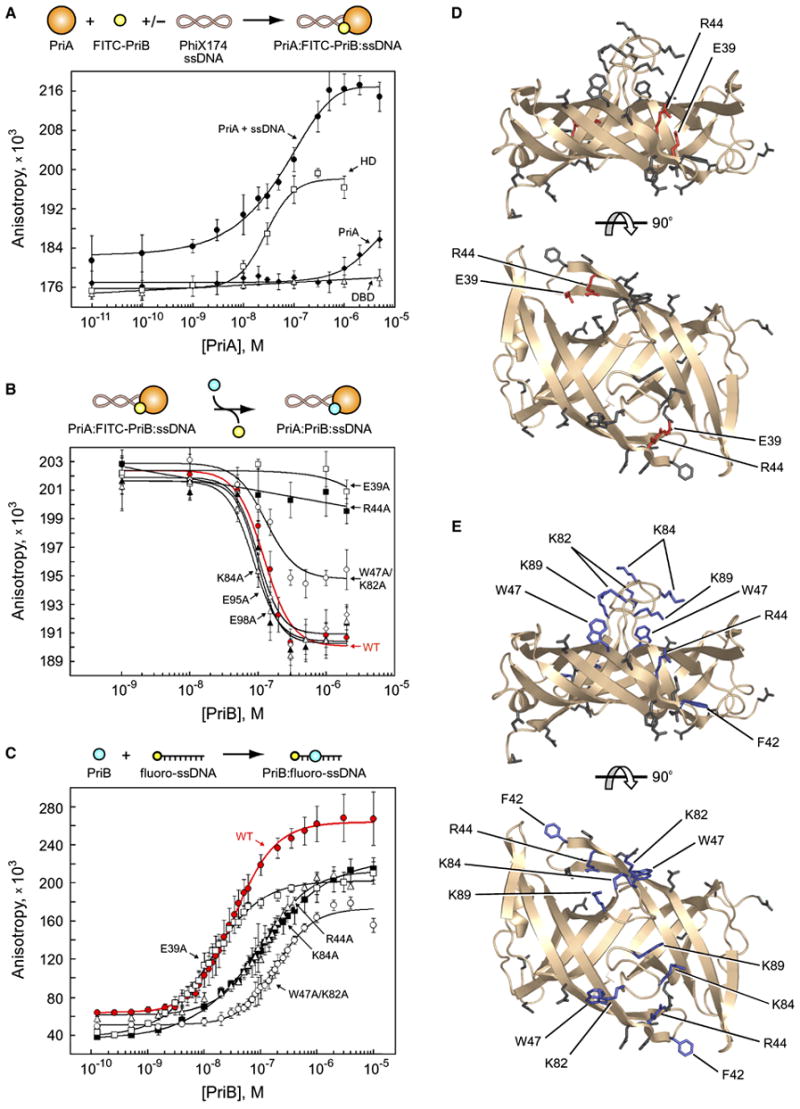
(A) Intact PriA (closed diamonds), PriA DBD (open triangles), PriA HD (open squares), and intact PriA in the presence of PhiX174 virion ssDNA (closed circles) were serially diluted and incubated with FITC-PriB. (B) Wild type PriB (red closed circles), E39A (open squares), W47A/K82A (open circles), R44A (closed squares), E95A (open diamonds), E98A (closed triangles), and K84A (open triangles) were serially diluted and incubated with PriA, FITC-PriB, and PhiX174 virion ssDNA. (C) Wild type PriB (red closed circles), E39A (open squares), R44A (closed squares), K84A (open triangles), and K89A (open circles) were serially diluted and incubated with fluorescein-labeled ssDNA oligonucleotide. Data are reported in triplicate and error bars are one standard deviation of the mean. For the sake of clarity, not all variants that were tested are shown. (D) Orthogonal views of ribbon diagrams of PriB show residues that are important for the PriA:PriB interface of the PriA:PriB:DNA ternary complex (red). Residues that were mutated but have no effect on the PriA:PriB:DNA ternary complex are shown in gray. The top view is the same as in Figure 1. (E) Orthogonal views of ribbon diagrams of PriB show residues that are important for ssDNA binding (blue). Residues that were mutated but have no effect on ssDNA binding are shown in gray.
