Figure 5. PriA:PriB:DnaT ternary complex formation and DnaB loading.
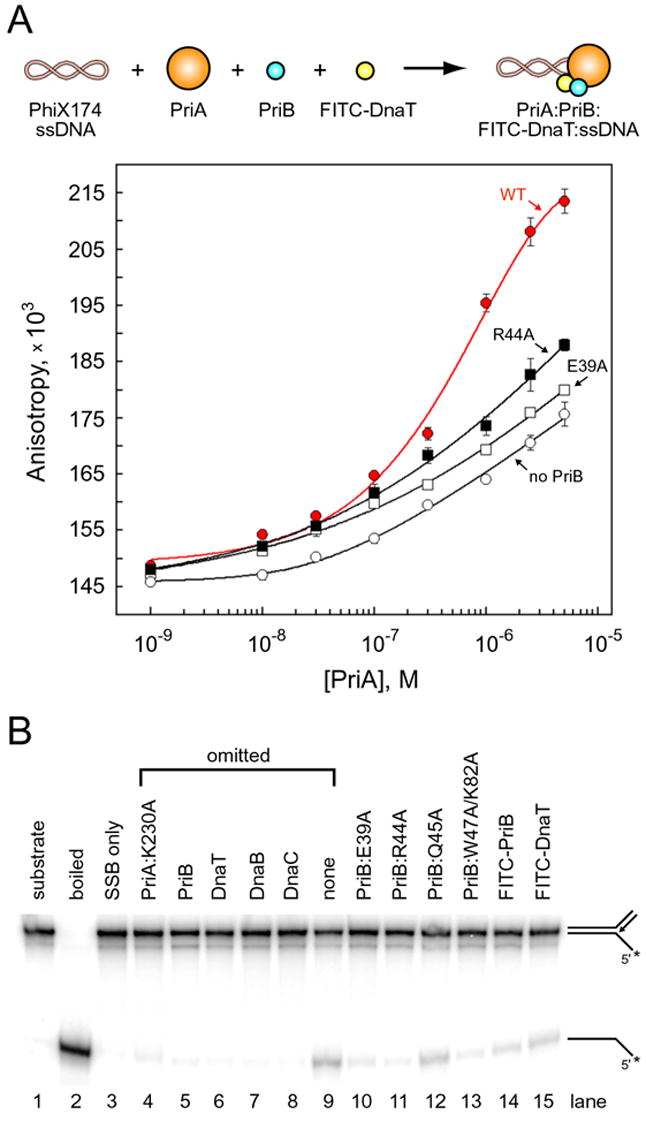
(A) PriA was serially diluted and incubated with PhiX174 virion ssDNA, FITC-DnaT, and either wild type PriB (red, closed circles), E39A (open squares), R44A (closed squares), or no PriB (open circles). Data are reported in triplicate and error bars are one standard deviation of the mean. (B) Lane 1, intact substrate (a fork with 60 bp of duplex DNA, a 38 nucleotide lagging strand arm, and a 38 bp duplex DNA leading strand arm); lane 2, boiled substrate; lane 3, substrate incubated with SSB only. The indicated proteins were omitted from reaction mixtures (lanes 4–9). PriB and DnaT variants were substituted for wild type PriB and DnaT, respectively (lanes 10–15). The PriA:K230A variant was used because it lacks ATPase and helicase activity, therefore any observed DNA unwinding results from DnaB activity. The positions of substrate and product are shown to the right of the gel and the position of the 32P label is indicated with an asterix.
