Fig. 1.
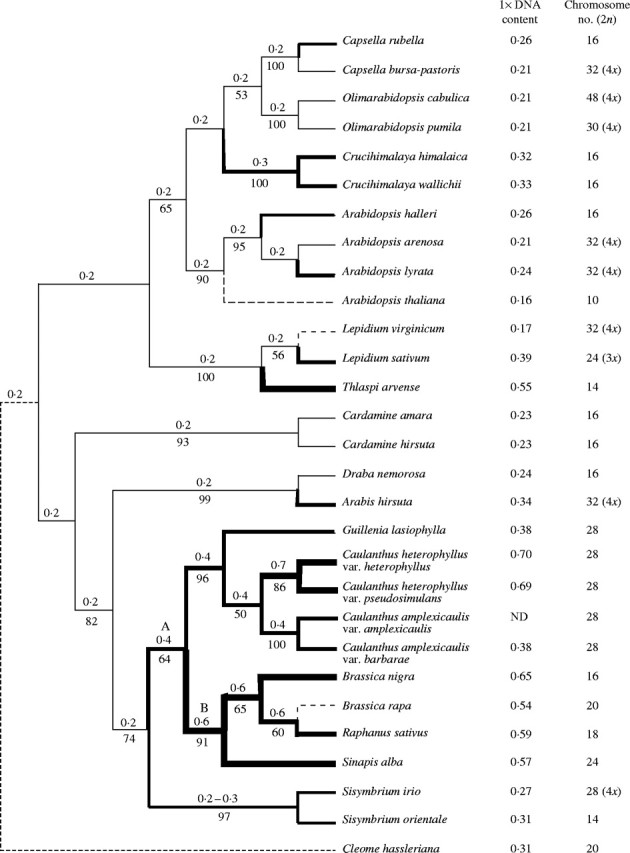
Phylogenetic analysis of the evolution of genome size in representative members of Brassicaceae based on a single most-parsimonious ITS tree. Bootstrap support (%) for various nodes (1000 replicates) is indicated beneath the corresponding node. Nodes without an indicated bootstrap support had bootstrap values of less than 50 %. Genome size (1× DNA content) expressed in pg, and chromosome numbers for each taxon are indicated. Numbers above each node indicate a hypothetical ancestral genome size (pg). Thickness of the lines shown in the tree is used to illustrate hypothetical changes in genome size in the Brassicaceae family. A dashed line is used to indicate a decrease in genome size. Cleome hassleriana (Capperaceae) merely provides an outgroup for the phylogenetic analysis, and was not used to estimate the ancestral genome size of Brassicaceae. A and B indicate nodes discussed in the text.
