FIG. 1.
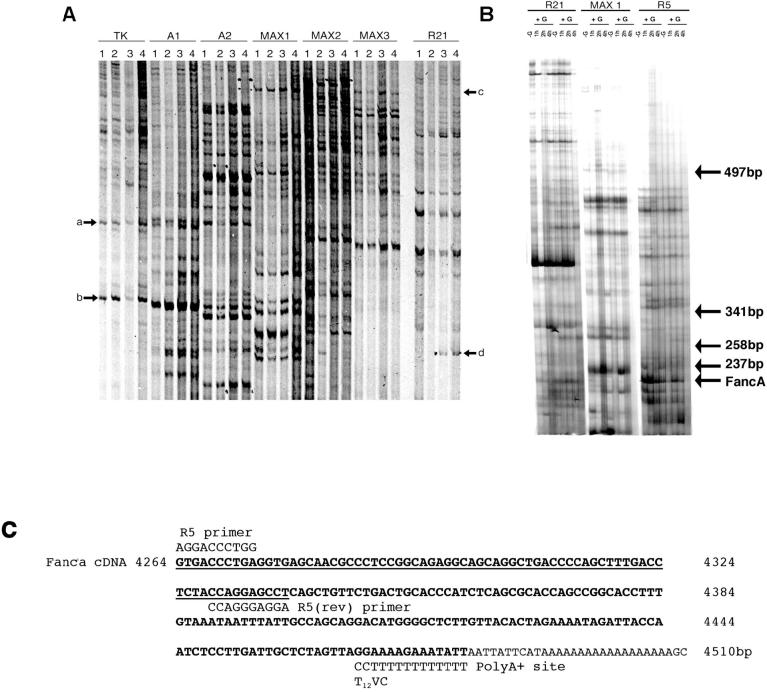
Differential display RT-PCR analysis of GnRH-regulated transcripts isolated from LβT2 cells. A) Matrigel basement membrane was excluded as an inducer of gene expression in LβT2 gonadotroph cells while GnRH was shown to upregulate transcripts. Cells were grown without Matrigel (1) or were cultured on Matrigel (2, 3, and 4) and treated with 3 (3) or 6 (4) 15-min pulses of GnRH with an interpulse interval of 75 min. RNA was extracted and subjected to differential display (DD) RT-PCR using primers stated. Matrigel did not alter transcript expression (arrows a and b), while numerous transcripts altered after GnRH treatment (arrows c and d). B) In two separate experiments, LβT2 cells were left untreated (−G) or were treated with 1 × 15-min pulse of GnRH (+G), RNA was harvested (−G, 1, 2, and 4 h) and subjected to DD-RT-PCR. First-strand cDNA was generated using the T12VC downstream primer, then amplified by PCR using primers R21, MAX1, or R5. Arrow denotes location of Fanca. The location of DNA size markers, indicated as bp, are also shown. C) Bioinformatic line-up depicting the region of homology, shown underlined, between the cloned Fanca DD-RT-PCR product and mouse Fanca cDNA nucleotide sequence, and the likely internal R5 priming site. The location of DD-RT-PCR primers R5 and T12VC that generated the original 216-bp DD-RT-PCR product are also indicated.