FIG. 3.
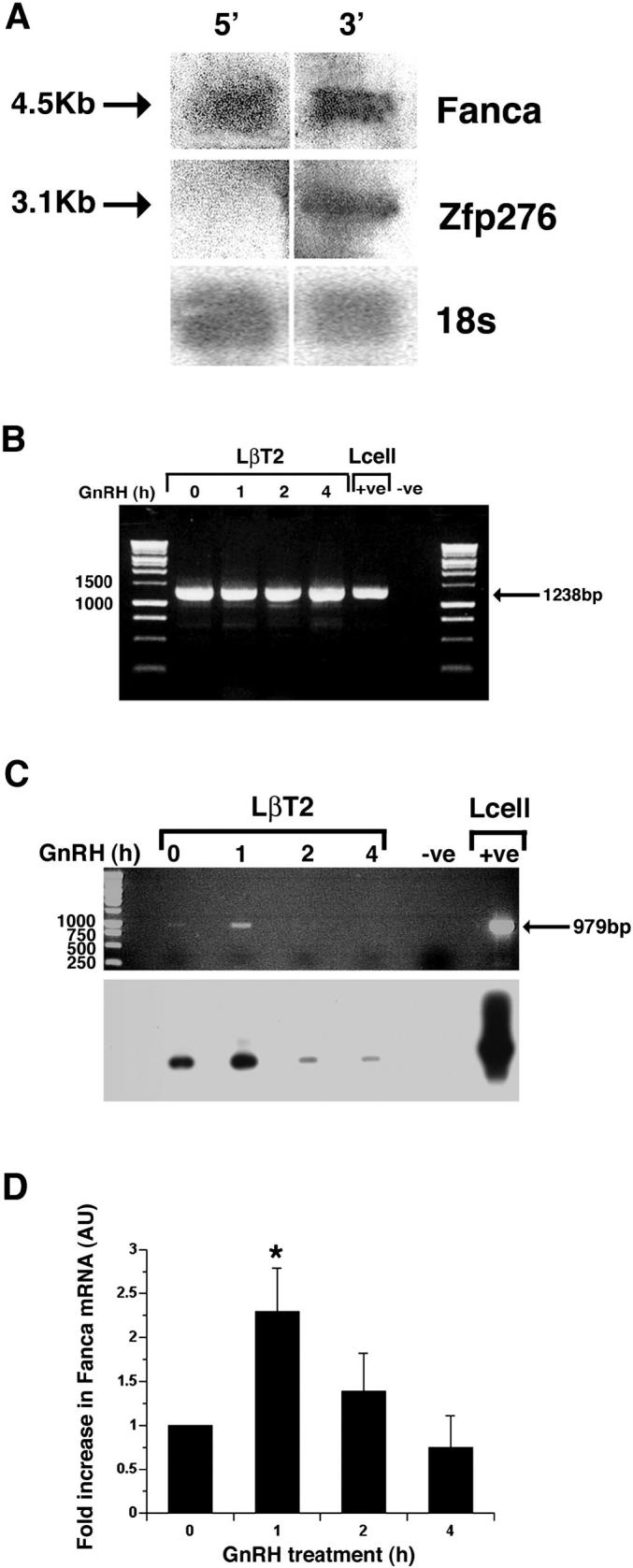
Characterization and quantification of Fanca and Zfp276 mRNA after GnRH treatment. A) Confirmation of expression of full-length 4.5-kb Fanca and 3.1-kb Zfp276 mRNA transcripts in LβT2 cells. Total RNA from LβT2 cells was fractionated on a formaldehyde gel, Northern blotted, and probed with radiolabeled probes corresponding to either the 5′ region (exons 1-11) or 3′ region (exons 42-43) of Fanca. The blot was then stripped and reprobed with an 18s probe. Specific Fanca, Zfp276, and 18s bands are indicated by arrows. B) Semiquantitative RT-PCR analysis of Zfp276 expression. LβT2 cells were left untreated (0) or treated with 1 × 15-min pulse of GnRH, then harvested 1, 2, and 4 h later, RNA was extracted, and first-strand cDNA made. PCR was performed to amplify full-length Zfp276 (1238 bp) and the products were visualized on an ethidium bromide-stained agarose gel. One specific 1238-bp PCR product was visible at all time points. L-cell cDNA was included as a positive control. An arrow denotes the 1238-bp PCR product, and DNA size markers are labeled. C) Semiquantitative RT-PCR analysis was performed for Fanca expression in LβT2 cells that were left untreated (0) or treated with 1 pulse of GnRH; harvested 1, 2, and 4 h later; RNA extracted, and cDNA made. Ethidium bromide staining identified one PCR product amplified from mRNA harvested from the 1-h time point. L-cell cDNA was included as a positive control. An arrow denotes the 979-bp PCR product, and size markers are labeled. Southern blotting analysis of the agarose gel with a 5′ Fanca probe confirmed that the Fanca PCR product was specific and present at all time points. D) LightCycler quantitative RT-PCR analysis of Fanca mRNA extracted from LβT2 cells either left untreated or treated with GnRH and harvested 1, 2, and 4 h later detected a consistent 2-fold increase in Fanca mRNA harvested 1 h after treatment. Results are shown as arbitrary units (AU) of Fanca mRNA normalized against the levels of internal control B-2-microglobulin (B2m) mRNA. This experiment was performed in duplicate and repeated three times. *P < 0.05 was determined as being significant by ANOVA one-way analysis of variance.
