Abstract
Mutations in MECP2 and Mecp2 (encoding methyl-CpG binding protein 2 [MeCP2]) cause distinct neurological phenotypes in humans and mice, respectively, but the molecular pathology is unclear. Recent literature claimed that the developmental homeobox gene DLX5 is imprinted and that its imprinting status is modulated by MeCP2, leading to biallelic expression in Rett syndrome and twofold overexpression of Dlx5 and Dlx6 in Mecp2-null mice. The conclusion that DLX5 is a direct target of MeCP2 has implications for research on the molecular bases of Rett syndrome, autism, and genomic imprinting. Attempting to replicate the reported data, we evaluated allele-specific expression of DLX5 and DLX6 in mouse × human somatic cell hybrids, lymphoblastoid cell lines, and frontal cortex from controls and individuals with MECP2 mutations. We identified novel single-nucleotide polymorphisms in DLX5 and DLX6, enabling the first imprinting studies of DLX6. We found that DLX5 and DLX6 are biallelically expressed in somatic cell hybrids and in human cell lines and brain, with no differences between affected and control samples. We also determined expression levels of Dlx5 and Dlx6 in forebrain from seven male Mecp2-mutant mice and eight wild-type littermates by real-time quantitative reverse-transcriptase polymerase chain reaction assays. Expression of Dlx5 and Dlx6, as well as of the imprinted gene Peg3, in mouse forebrain was highly variable, with no consistent differences between Mecp2-null mutants and controls. We conclude that DLX5 and DLX6 are not imprinted in humans and are not likely to be direct targets of MeCP2 modulation. In contrast, the imprinting status of PEG3 and PEG10 is maintained in MeCP2-deficient tissues. Our results confirm that MeCP2 plays no role in the maintenance of genomic imprinting and add PEG3 and PEG10 to the list of studied imprinted genes.
Rett syndrome (RTT [MIM #312750]) is a neurodevelopmental disorder that affects females almost exclusively. An apparently normal early-postnatal period is followed by developmental stagnation and then regression, with loss of motor skills and speech, autonomic dysfunction, and seizures.1 De novo recurrent loss-of-function mutations of the X-linked gene MECP2 (MIM +300005) are found in almost all girls who receive a clinical diagnosis of RTT2 (for review, see the work of Francke3). The incidence is estimated to be 1 in 10,000 female births, and the availability of molecular diagnostic testing leads to earlier diagnosis, before the clinical picture is fully developed. MECP2 encodes methyl-CpG binding protein 2 (MeCP2), a multifunctional protein that is expressed ubiquitously but at the highest levels in neurons. Since MECP2 is subject to X inactivation, affected females are mosaic for cells that either have normal MeCP2 levels or lack MeCP2 function completely. Classic RTT is associated with random X-inactivation patterns, but skewed X-chromosome inactivation leads to phenotypic variants.4,5 Most males with inactivating MECP2 mutations have congenital encephalopathy, with lack of postnatal development, and respiratory insufficiency that usually leads to early death.6 Recent elegant experiments with genetically manipulated mouse models revealed that the symptoms can be prevented or delayed7 and even reversed8 when normal MeCP2 function is restored postnatally.
One of the functions of MeCP2 is to repress transcription of methylated genes by recruiting a chromatin remodeling complex to promoter regions.9 Therefore, lack of MeCP2 is expected to cause abnormal expression of genes that affect postnatal neuronal function. Although large-scale misregulation of gene expression has not been observed, a few target genes have been identified by global gene-expression studies.10–17 Imprinted genes that are transcribed exclusively from the maternal or the paternal allele are attractive candidates for potential MeCP2 modulation, because their uniparental expression pattern is controlled by parent-of-origin–specific methylation of specific sites (differentially methylated regions [DMRs]).18 Of the ∼80 known imprinted genes in mammals,19 5 (SNRPN, IPW, NDN, H19, and IGF2) were previously studied in RTT tissues. These genes showed monoallelic expression in MeCP2-deficient clonal cell lines and brain.20 Although Samaco et al.21 and Makedonski et al.22 reported decreased expression levels of the brain-imprinted genes UBE3A and Ube3a in MeCP2-deficient human and mouse brain tissues, respectively, these data could not be replicated in a more extensive study of Ube3a RNA and protein expression in Mecp2-mutant mouse brains.23
Hailed as a breakthrough discovery with a major impact on RTT research was the work by Horike et al.24 that reported that MeCP2 regulates the expression of the distal-less homeobox 5 gene (DLX5). Under the assumption that DLX5 is imprinted, the authors claimed loss of imprinting in MeCP2-deficient cell lines and brain. In an Mecp2-mutant mouse model,25 expression levels of Dlx5 and Dlx6 were reported to be increased twofold in the frontal cortex. This increase was said to be the result of “relaxed imprinting,” although Dlx5 is known to be biallelically expressed in mice.26 Horike et al.24 concluded that DLX5 is a direct target of MeCP2 modulation, and this conclusion is now widely quoted in the literature.
Using a wide range of methodologies, Horike et al.24 reported a series of observations that are not coherent. First, searching for in vivo binding sites of MeCP2, they precipitated urea-gradient–purified, formaldehyde–cross-linked chromatin, derived from whole brains of 1-d-old normal mice, with anti-MeCP2 antibody and cloned the precipitated DNA fragments. Of 100 randomly sequenced clones, only 3 contained CpG dinucleotides and were consistent with promoters. The others were located in introns or up to 100 kb away from the ends of the nearest transcription unit. Scanning the vicinity of apparent MeCP2-binding sequences for biologically interesting candidates, Horike et al.24 focused on DLX5 for two reasons. First, DLX5 directly regulates expression of glutamic acid dehydroxylase and promotes differentiation of GABAergic neurons,27 and, second, DLX5 had previously been reported to be imprinted in human lymphoblasts and brain tissues,28 although not in mouse brain.26
DLX genes encode a family of transcription factors that contain a homeobox DNA-binding domain related to that of the Drosophila gene distal-less.29 In mammalian genomes, the six known DLX genes occur in pairs that are closely linked. DLX5 and DLX6 are located within a 20-kb region in a tail-to-tail configuration on human chromosome 7q21.3 and mouse chromosome 6A1. During development, DLX5 and DLX6 are expressed in defined regions of the brain and in skeletal structures. DLX5 induces bone formation and is expressed in later stages of osteoblast differentiation. Dlx5-knockout mice have multiple defects in their ears, noses, mandibles, and skull bones and die shortly after birth.30 A proportion of them have exencephaly.31 Early developmental defects in neurogenesis have also been recognized.32 Notably, developmental defects were seen only in homozygous mutants, not in heterozygotes with a parent-of-origin–dependent effect, as would be expected if Dlx5 were an imprinted gene.
Evidence that DLX5 is imprinted in humans was first reported by Okita et al.,28 who studied somatic cell hybrid (SCH) lines containing a single human chromosome 7 of defined parental origin. DLX5 was not expressed in SCHs containing a paternal chromosome 7 but was expressed in cells containing a maternally derived chromosome 7. The hybrid cell studies of 76 ESTs from the 7q21-q31 region revealed monoallelic expression for six transcripts, but DLX5 was the only one for which imprinting status could be confirmed by studies of human lymphoblastoid cell lines (LCLs) and brain. Specifically, Okita et al.28 studied LCLs from 15 individuals who were heterozygous for an intragenic polymorphism in DLX5 (c.*163dupC [rs5886002]; the alleles are referred to as “-/G” in dbSNP). RT-PCR products were sequenced and were said to reveal monoallelic expression in 13 of the 15 samples. In all eight cases in which the parental origin of the alleles could be determined, DLX5 was reported to be expressed only from the maternal allele. The authors also examined the allelic expression of DLX5 in brain tissue samples from three c.*163dupC heterozygotes. Sequence analysis of RT-PCR products revealed biallelic but unequal expression, preferentially from a single allele of unknown parental origin. Nevertheless, Okita et al. concluded that “DLX5 is imprinted and maternally expressed in normal human lymphoblasts and brain tissues.”28(p557)
Horike et al.24 used the same c.*163dupC polymorphism to assess the imprinting status of DLX5 in LCLs from individuals with RTT and controls. By using RT-PCR with 40 cycles, they reported monoallelic expression in two normal control LCLs and in only one of four RTT LCLs. They interpreted these results to indicate “loss of imprinting” in RTT.
In the mouse, Kimura et al.26 reported that Dlx5 is not imprinted. They studied offspring of interspecies crosses, distinguished by a SNP in the 3′ UTR of Dlx5, as well as heterozygous Dlx5-knockout mice that had inherited the knockout allele from either parent. Both approaches revealed that Dlx5 is expressed in a biallelic fashion in cerebral cortex, diencephalon, olfactory bulb, hippocampus, and testis, with no allele-specific preference. Furthermore, the CpG islands of the Dlx5 and Dlx6 promoters were unmethylated on both alleles in wild-type and Mecp2 Y/- mice.24 Moreover, no differential methylation was detected at other CpG sites in the Dlx5/Dlx6 region between the two alleles, either in wild-type or in Mecp2-mutant mice. The absence of DMRs is consistent with the observed lack of imprinting of Dlx5 and Dlx6 in mouse brain.
Horike et al.24 assessed expression of Dlx5 and Dlx6 in mouse brain, first by quantitative RT-PCR (qRT-PCR) experiments on brain samples from an unspecified number of 8-wk-old wild-type and Mecp2-mutant mice. When relative expression levels for Dlx5 and Dlx6, as well as for other imprinted and nonimprinted genes, were compared, only Dlx5 and Dlx6 showed an approximately twofold increase in the mutant samples compared with in the wild-type samples. To determine whether the increased expression was allele specific, Horike et al. examined the SNP in the 3′ UTR of Dlx5 in interspecies crosses, as was done previously by Kimura et al.26 On the basis of the intensity of restriction-enzyme fragments of RT-PCR products, Horike et al. confirmed that Dlx5 was biallelically transcribed in frontal cortex, but transcript levels were said to be higher for the maternal allele. However, in an Mecp2-mutant male mouse, which was heterozygous for the Dlx5 SNP, both parental-specific restriction fragments were present at equal levels, which led the authors to conclude that MeCP2 deficiency abolished the albeit incomplete imprinting pattern. Notably, the transcript levels and parental-specific expressions of four other, truly imprinted genes (Calcr, Sgce, Peg10, and Asb4), located in the same gene cluster on chromosome 6, were not affected by MeCP2 deficiency.
To examine the validity of the fundamental claims that led to the identification of DLX5 as a primary target of MeCP2, we attempted to reproduce the reported data in a systematic fashion. First, in two sets of SCHs with single paternal or maternal copies of chromosome 7, we found expression of both alleles of human DLX5 and DLX6. Second, both genes were consistently expressed in a biallelic fashion in LCLs and brain samples from normal individuals and in clonal RTT LCLs. To evaluate whether MeCP2 deficiency relaxes the imprinting of truly imprinted genes in humans, we studied allele-specific expression of Paternally expressed gene 3 (PEG3) on chromosome 19 and Paternally expressed gene 10 (PEG10) near the DLX5 and DLX6 genes on 7q. We demonstrated strictly monoallelic expression of PEG3 and PEG10 in tissues from males and females with MECP2 mutations. Third, to evaluate the alternative hypothesis—also put forward by Horike et al.,24 that differential chromatin loop formation affecting the expression of both alleles, rather than allele-specific imprinting, could control Dlx5 transcript levels in different brain regions—we compared expression levels of Dlx5 and Dlx6 in the forebrain of Mecp2-mutant male mice and normal male littermates. We saw no differences when we combined data from seven different litters of mice aged 7–9 wk. Our results provide strong evidence against the claim that DLX5 and DLX6 are targets of MeCP2 and play a role in the pathogenesis of RTT.
Material and Methods
SCHs
Human blood leukocytes from unaffected donors were fused with the Hprt-deleted mouse cell line A9. SCHs were selected in hypoxanthine-aminopterin-thymidine medium, which forces retention of the human X chromosome. Hybrid cell clones were genotyped for eight microsatellite markers on human chromosome 7, to identify those that had retained a single copy of chromosome 733 (performed by GMP Genetics). Genotyping of the donors’ parents for the loci with distinguishing alleles allowed us to assign maternal or paternal origin of the chromosome 7 that was present. Chromosome 7 retention was not selected for in the hybrid clones, and we did not determine the copy numbers. Hybrid cell lines were grown in Dulbecco’s modified Eagle medium with 10% fetal calf serum, 2 mM glutamine, and 1× hypoxanthine and thymidine and were subcultured at 1:3 with TrypLE (GIBCO), by use of standard tissue-culture techniques.
LCLs and Brain Samples
For detection and genotyping of DLX5 and DLX6 polymorphisms, we sequenced 14 LCLs previously established in our laboratory from RTT-affected individuals with known MECP2 mutations, 22 unaffected control LCLs, and six fetal and six adult control brain samples. For DLX5, 15 additional control brain samples were genotyped. For two PEG3 SNPs, we genotyped the RTT LCLs and seven brain samples from individuals with RTT, including two hemizygous males with known MECP2 mutations (University of Maryland Brain and Tissue Bank for Developmental Disorders [numbers 1238, 1420, 1748, 1815, 4516, and 4852]). One sample from the Harvard tissue bank is brain sample number b4160. Only the brain samples were genotyped for PEG10 SNPs. Frozen tissue used for the expression studies was from frontal cortex. All human materials were obtained and studied under a protocol approved by the Stanford Human Research Protection Program.
DNA Extraction and Genotyping
We extracted DNA from cultured cells, by using phenol-chloroform, and from brain tissue, by using DNA Stat 60 (Tel-Test). For PCR, we used Promega Go Taq in accordance with the manufacturer’s instructions, with the primers listed in table 1, in a 25-μl reaction. The PCR cycling program included an initial denaturation at 95°C for 4 min, followed by five cycles at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. The annealing temperature was reduced by 0.5°C in the second to fifth cycle. For 30 additional cycles, the annealing temperature was 54°C for the DLX5 genomic DNA (gDNA) primers and was 57°C for the DLX5 cDNA primers, as well as for the DLX6, PEG3, and PEG10 primers. The gel-purified PCR amplicons were sequenced using BigDye Terminator chemistry on an ABI 3100 sequencer (Applied Biosystems). For confirmatory studies of the DLX5 c.*163dupC polymorphism, we used a proofreading DNA polymerase (Platinum Pfx [Invitrogen]) to minimize slippage errors during the amplification of the mononucleotide strings.
Table 1. .
Primers for Genotyping and Expression Analysis
| gDNA Primer(5′→3′) |
cDNA Primer(5′→3′) |
||||||
| Organism and Gene/SNP | Alleles | Forward | Reverse | Product Size (bp) |
Forward | Reverse | Product Size (bp) |
| Human: | |||||||
| DLX5 c.*163dupC (rs5886002) | -/G | TTTTTTGGGACTACTGTGTTTTGC | AGATTTCAAGGCACCATTGAAAG | 203 | GCTGGGATTGACACAAACAC | AGGCACCATTGAAAGTGTCC | 568 |
| DLX5 c.*142T→C | T/C | Same as above | Same as above | Same as above | Same as above | ||
| DLX5 5′ UTR | … | … | … | … | GCCACAACAGCAAGGACAG | TTTGCCATTCACCATTCTCA | 440 |
| DLX6 c.*9A→G (rs3213654) | A/G | CTCCAGTCTGGGACGTTTCT | GCTCTCCTAAGCCTGCTCCT | 232 | TCGCTTTCAGCAGACACAGT | CGGCTTCTTGCCACACTTAT | 457 |
| DLX6 c.*775dupC | -/C | AAGGGAATGCTGCATGTTTT | TAGCTTTGTGAATGCCACCA | 202 | Same as for gDNA | Same as for gDNA | … |
| DLX6 c.*771C→T (rs2272280) | C/T | Same as above | Same as above | Same as above | Same as above | ||
| PEG3 c.*703A→G (rs1055359) | A/G | CTTGTGAAGCTGTAGGCATGA | CTGGGTCACAAAAAGCCAAT | 163 | Same as for gDNA | Same as for gDNA | … |
| PEG3 c.42C→T (rs1860565) | C/T | GGGATGGGTACTCACCACTG | CAGGTCATTCCAACCATGTG | 218 | AAGCCGGAGAACTGTGAGAA | CTTCTTGGGTTCCTGGTGTG | 239 |
| PEG10 c.*2923T→C (rs13073) | C/T | GTGTCATTTTCCTGCCTGGT | AGGAGCCTCTCATTCACAGC | 410 | Same as for gDNA | Same as for gDNA | |
| Mouse: | |||||||
| Dlx5 | … | … | … | … | TCTCTAGGACTGACGCAAACA | GTTACACGCCATAGGGTCGC | 132 |
| Dlx6 | … | … | … | … | TTCCCGAGAGAGCCGAACT | GTGGGTTACTACCCTGCTTCA | 117 |
| Peg3A | … | … | … | … | CACGAAGACGACACCAACAG | GTCTCGAGGCTCCACATCTC | 150 |
| Peg3B | … | … | … | … | ACAGTGACGACGACATGAGC | GTCTCGAGGCTCCACATCTC | 122 |
| Rps28 | … | … | … | … | TAGGGTAACCAAAGTGCTGGGCAG | GACATTTCGGATGATAGAGCGG | 103 |
| β-actin | … | … | … | … | TGACCCTGAAGTACCCCATTGA | CCATGTCGTCCCAGTTGGTAAC | 54 |
| Snca | … | … | … | … | GCAGCCAGAAGTCGGAAA | TGAACACATCCATGGCTAAAGA | 58 |
RNA Extraction, cDNA Synthesis, and RT-PCR Assay
We extracted total RNA from mouse × human SCH lines and frozen human cortex, using RNA Stat 60 (Tel-Test). From the LCLs, we extracted mRNA by use of Oligotex Direct mRNA Mini Kit (Qiagen). We treated the RNA with 20 U RNase-free DNaseI (Roche) for 20 min, followed by 10 min of inactivation of the enzyme at 75°C. For reverse transcription, 5 μg total RNA or 1 μg mRNA was incubated with Superscript III (Invitrogen), as recommend by the manufacturer, in a 40-μl reaction. For each reaction, we used a “minus RT” control, to which no Superscript III was added. For DLX5, DLX6, PEG3, and PEG10 RT-PCR assays, we used gene-specific primers (table 1) and 4 μl of the cDNA reaction with Promega Go Taq Flexi, in accordance with the manufacturer’s instructions. PCR cycling and sequencing of amplicons were done as described above.
Mecp2-Mutant Mice
Female mice heterozygous for the Mecp2tm1.1Bird mutation25 were purchased from the Jackson Laboratory and were bred with C57BL/6J male mice. Tail DNA samples were genotyped for Mecp2, as described elsewhere.23 Mutant male mice and wild-type male littermates at age 7–9 wk were euthanized with CO2. Forebrains (the frontal one-third of the cortex) were dissected and were snap frozen on dry ice. The Animal Care Committee of Stanford University approved all experimental procedures.
Real-Time qRT-PCR of Dlx5, Dlx6, and Peg3 in Mouse Brain
We extracted total RNA from frozen brain tissues by using TRIzol reagent (Invitrogen). We treated 2 μg of total RNA with 2 U DNase I (Ambion) and reverse transcribed it with random hexamers and Superscript II (Invitrogen). After real-time qRT-PCR with Sybr Green on an ABI 5700 instrument (AME Bioscience), we analyzed melting curves for each reaction, to ensure a single peak. Actb and Rps28 served as RNA controls for relative quantitation (User Bulletin 2 [Applied Biosystems]). Details of the methods were as described in previous articles from our laboratory.12,34 Primer sequences are listed in table 1. Dlx5 primers (nt 617–636 and nt 748–729 of GenBank accession number NM_198854.1) span the last two exons—that is, exons 2 and 3 or exons 3 and 4, depending on the transcript variant. Dlx6 primers (nt 339–319 and nt 223–241 of GenBank accession number BC114342.1) span exons 2 and 3. For Peg3, two different primer sets were used: Peg3A, spanning exons 2, 3, and 4, and Peg3B, spanning exons 3 and 4.
Results
Expression of Human DLX5 and DLX6 from Maternal and Paternal Chromosome 7 in Mouse × Human SCH Lines
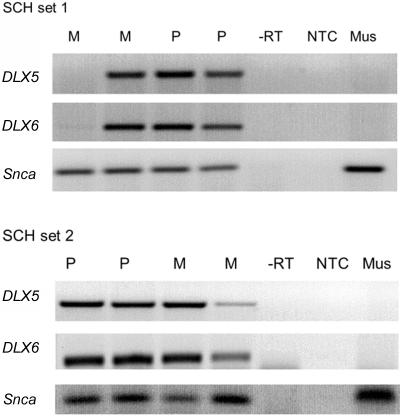
We examined eight SCH lines, four each derived from two unrelated individuals. In both sets, two lines had retained the maternal copy of chromosome 7 and two had retained the paternal copy, as determined by STR genotyping. DLX5 and DLX6 were consistently expressed from both alleles in seven of the eight SCH clones (fig. 1). Quantitative expression differences between clones are likely caused by different retention frequencies of chromosome 7 in the SCH lines, and the one exceptional clone with no transcript had apparently lost chromosome 7 during propagation of the culture. For a loading control, we studied mouse Snca expression. The DLX5 primers specifically amplify the human transcript, including the 5′ UTR, exon 1, and part of exon 2. The forward primer is complementary only to human DLX5, and the reverse primer has seven mismatches with the mouse Dlx5 sequence. The two DLX6 primers from the 3′ UTR had four and two mismatches with the Dlx6 sequence. Both sets of primers did not amplify mouse cDNA, since no transcripts were obtained from the SCH without chromosome 7 or from mouse control brain. When we sequenced DLX5 and DLX6 RT-PCR products from SCH lines, both were 100% identical to the human cDNA sequences. Thus, our results provide clear evidence of biallelic expression of DLX5 and DLX6 from both parent-specific copies of human chromosome 7 retained in mouse × human SCHs. Our data contradict a previous report of maternal-specific imprinted expression of DLX5.28 The imprinting status of DLX6 had not been determined previously.
Figure 1. .
Absence of imprinting of DLX5 and DLX6 in SCH lines. RT-PCR analysis of human DLX5 and DLX6 expression in mouse × human SCHs containing a single copy of human chromosome 7 shows that both genes are expressed from the maternally (M) as well as the paternally (P) derived chromosome 7. One SCH line in set 1 had apparently lost the maternal chromosome 7, and one line in set 2 had retained the maternal chromosome 7 at a low copy number. Primers used do not amplify the mouse (Mus) orthologues in C57BL/6 cortex. Mouse Snca expression is shown as a loading control. NTC = no template control; -RT= first-strand synthesis without reverse transcriptase.
Biallelic Expression of DLX5 in RTT LCLs, Normal LCLs, and Normal Human Brain
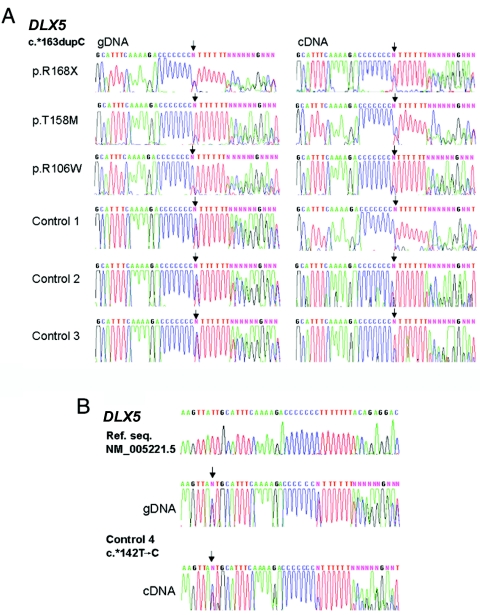
Next, we tested expression and imprinting status of DLX5 in LCLs and brain samples by studying the same c.*163dupC polymorphism that previously was used to distinguish the two alleles24,28 (fig. 2A). To identify heterozygotes, we sequenced gDNA from 36 human LCLs and 27 brain samples. We confirmed the results with a second round of sequencing, using a proofreading polymerase. Three LCLs initially designated as heterozygous were found to be C7 homozygotes after resequencing. Frequencies of c.*163dupC heterozygotes were 5/14 for RTT LCLs, 6/22 for normal LCLs, and 6/27 for normal brains. A novel SNP, c.*142T→C, was discovered in one sample (fig. 2B). Allele frequencies are provided in table 2.
Figure 2. .
Absence of imprinting of DLX5 in LCLs. A, Biallelic expression revealed by DLX5 genotyping (of gDNA) and expression analysis (of cDNA) of LCLs from three females with RTT and common MECP2 mutations, as indicated, and three normal controls. All six LCLs are heterozygous for the DLX5 c.*163dupC polymorphism (rs5886002) (left panel), and all show that both alleles are expressed (right panel). The sequence shown is the sense strand. Arrows indicate the positions of nucleotide changes. B, Novel DLX5 SNP c.*142T→C detected in a normal LCL, denoted by an arrow in the gDNA. Comparison with the reference sequence (Ref. seq. [GenBank accession number NM_005221.5) shows that the SNP is located upstream of rs5886002. LCLs from control 4 are heterozygous for both SNPs, and both alleles are expressed in cDNA.
Table 2. .
DLX5 and DLX6 Sequence Variants[Note]
| SNP | dbSNP | No. of Unrelated Individuals Studied | Allele Frequencies | |
| DLX5 c.*163dupC | rs5886002 | 64 | - = .87 | C = .13 |
| DLX5 c.*142T→C | No record | 50 | T = .99 | C = .1 |
| DLX6 c.*9A→G | rs3213654 | 46 | A = .98 | G = .2 |
| DLX6 c.*775dupC | No record | 50 | - = .99 | C = .1 |
| DLX6 c.*771C→T | rs2272280 | 50 | C = .98 | T = .2 |
Note.— Reference sequences were DLX5 (GenBank accession number NM_005221.5) and DLX6 (GenBank accession number NM_005222.2). All variants are described in accordance with guidelines from den Dunnen and Antonarakis35; updates can be found at HGVS Nomenclature for the Description of Sequence Variants.
By RT-PCR analysis, DLX5 was biallelically expressed in all 13 heterozygous samples that showed expression levels sufficiently high for analysis (figs. 2 and 3). The results are unlikely to be caused by gDNA contamination, because the primers used for amplification spanned an intron, and no PCR product was obtained in the absence of reverse transcriptase in the first-strand cDNA synthesis reaction. The sequence tracings suggest apparently equal amplification of transcripts from both alleles in most LCLs from individuals with RTT and unaffected controls, except for the sample with p.R106W and control 1, for which the cDNA results suggest preferential amplification of the C7 allele. Similar skewing is apparent, however, for the gDNA samples with p.T158M and p.R106W and likely is caused by PCR artifacts during amplification of the C7/C8 alleles (fig. 2). There was no obvious difference between the heterozygous clonal RTT LCLs that expressed either mutant MECP2 in 100% of cells (p.R168X and p.T158M) or wild-type MECP2 in 100% of cells (p.R106W).12
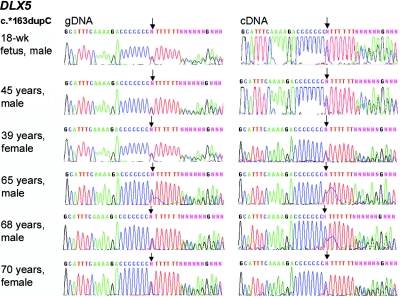
Figure 3. .
Absence of imprinting of DLX5 in frontal cortex. DLX5 genotyping (of gDNA) and expression analysis (of cDNA) of frontal cortex brain samples from unaffected controls revealed biallelic expression. All six donors (age and sex indicated) were heterozygous for the c.*163dupC (rs5886002) polymorphism (left panel). In all brain samples, both alleles are expressed at a similar level (right panel). Arrows indicate the positions of nucleotide changes.
The low expression levels of DLX5 and DLX6 in LCLs presented a considerable challenge. Initially, when we raised the PCR cycle number to ⩾40, we occasionally saw expression of only one or the other allele, but not in a parent-of-origin–specific manner. These results were not reproducible and were probably caused by the very low copy number of transcripts. We obtained consistent and reproducible biallelic expression only when we performed RNA extraction, first-strand cDNA synthesis, and PCR amplification in one session, avoiding freeze-thaw cycles of the samples. In contrast, RT-PCR of all six frontal cortex samples from non-RTT donors generated strong and unambiguous biallelic expression of DLX5 (fig. 3). We conclude that DLX5 is not imprinted in human LCLs and cortex.
Biallelic Expression of DLX6 in Normal Human Brain
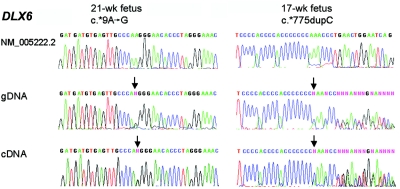
The imprinting status of DLX6 had not been determined previously because of a lack of suitable polymorphisms.24 We screened our LCLs and brain samples for heterozygosity at two rare SNPs (c.*9A→G [rs3213654] and c.*771C→T [rs2272280]) that are located ∼800 nt apart in the 3′ UTR and are in linkage disequilibrium. One of 22 normal LCLs and 1 of 12 non-RTT brain samples were heterozygous for both SNPs. In addition, one fetal brain gDNA showed a novel c.*775dupC variant. By RT-PCR, DLX6 expression in the LCLs was too low for reproducible results; however, we obtained reliable data for the two brain samples, and both showed biallelic expression of DLX6 (fig. 4).
Figure 4. .
Absence of imprinting of DLX6 in fetal brain. DLX6 genotyping (of gDNA) and expression analysis (of cDNA) in two fetal brain samples revealed biallelic expression. The 21-wk fetus (left panel) was heterozygous for the known c.*9A→G SNP (rs3213654). The 17-wk fetus (right panel) was heterozygous for a novel SNP, c.*775dupC. The upper trace displays the reference sequence, GenBank accession number NM_005222.2. Arrows indicate the positions of nucleotide changes.
Monoallelic Expression of Imprinted Genes PEG3 and PEG10 in MeCP2-Deficient Tissues
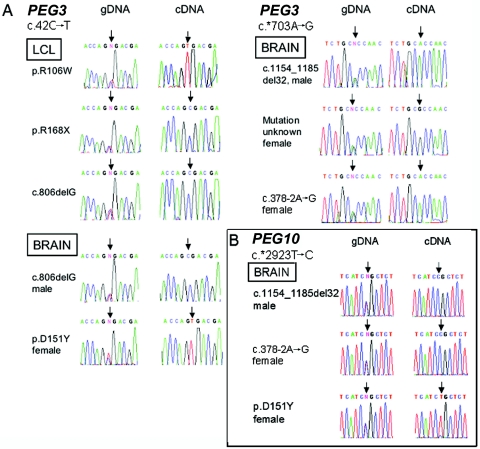
Because we were unable to confirm that DLX5 and DLX6 are imprinted in humans, our results could not address the question of whether MeCP2 deficiency leads to “relaxation of imprinting” in our samples. Therefore, we initially examined the bona fide imprinted gene PEG3 as a positive control. Of 13 RTT LCLs, 7 were heterozygous for the common c.42C→T polymorphism (rs1860565). We tested the allele-specific PEG3 expression of three LCLs carrying different MECP2 mutations (p.R106W, p.R168X, and c.806delG). By using the androgen-receptor assay and MECP2 expression analyses, we had previously discovered completely skewed X-inactivation patterns in these three LCLs.12 Although the p.R168X LCLs expressed the mutant MECP2 allele in 100% of cells, the p.R106W and c.806delG LCLs were 100% wild type for MECP2 expression. All three LCLs showed strictly monoallelic expression of PEG3 (fig. 5A), indicating that imprinting of this gene is maintained in the absence of MeCP2 protein.
Figure 5. .
Imprinting of PEG3 and PEG10 in MECP2 mutants. A, Maintenance of imprinting of PEG3 in cell lines and brains from males and females with MECP2 mutations. Samples on the left are heterozygous for the PEG3 c.42C→T SNP (rs1860565) (gDNA); samples on the right are heterozygous for the c.*703A→G SNP (rs1055359) (gDNA). Arrows indicate the locations of variant nucleotides. As shown in the right-hand traces (cDNA), all samples express PEG3 from only one allele. B, Maintenance of imprinting of PEG10 in frontal cortex from one male and two females with MECP2 mutations who are heterozygous for the c.*2923C→T SNP (rs13073). RT-PCR analyses reveal strictly monoallelic expression (cDNA). Arrows indicate the positions of variant nucleotides.
To assess the PEG3 imprinting status in brain, we tested five brain samples from females with RTT for two PEG3 SNPs, c.*703A→G (rs1055359) and c.42C→T (rs1860565), and identified three heterozygotes. One had no known MECP2 mutation, the second had missense mutation c.451G→T (p.D151Y), and the third was reported to have c.378-2A→G, affecting the 5′ splice site of exon 4. By RT-PCR analysis of this brain sample, we were able to document that this mutation leads to abnormal splicing, causing a frameshift and premature stop codon. The mutant and wild-type MECP2 transcripts were amplified to similar degrees (data not shown). We also included two brain samples from males with congenital encephalopathy who were hemizygous for c.806delG and c.1154_1185del32 MECP2 mutations and heterozygous for one of the PEG3 SNPs. All five brain samples showed strictly monoallelic expression of PEG3 (fig. 5A).
To extend the control studies to another imprinted gene, we selected PEG10 because it is located on chromosome 7 near the DLX5 and DLX6 loci. Three of our six MECP2-mutant brain samples genotyped for SNPs, one male and two female, were heterozygous for the common PEG10 SNP c.*2923T→C (rs13073). Only one PEG10 allele was expressed in each of the samples (fig. 5B). We conclude that the MeCP2 protein is not necessary for the maintenance of PEG3 and PEG10 imprinting in human brain.
Highly Variable Expression Levels of Dlx5, Dlx6, and Peg3 in Mouse Brain
Although Dlx5 is not imprinted in mouse, Horike et al.24 reported a twofold increase in expression of Dlx5 and Dlx6 in Mecp2-mutant brain compared with that in control brain. To explain that increase, they postulated that MeCP2 induces a specific “silent chromatin loop” formation that affects expression levels, presumably on both alleles. As the first step to evaluate the validity of this model, we attempted to reproduce the reported expression changes.
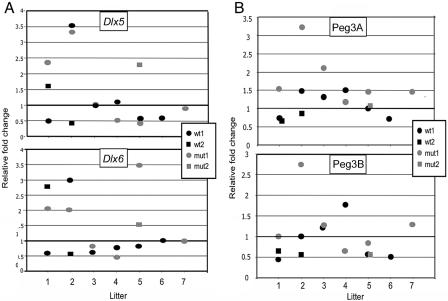
We established robust qRT-PCR assays on mouse forebrain in which Dlx5 and Dlx6 are highly expressed. Both the Dlx5 and Dlx6 primers span an intron, and the Dlx5 primers amplify both splice variants. We used mice with the same mutant allele (Mecp2tm1.1Bird) on the same strain background (C57BL/6J) and bred them with mice of the same age (7–9 wk) as in the previously published report. We normalized qRT-PCR data for Dlx5 and Dlx6 to two housekeeping genes whose expression is not affected by MeCP2 deficiency at that age: Actb (encoding β-actin) and Rps28.17 We then compared the relative transcript levels of Dlx5 and Dlx6 in 15 male mice (7 Mecp2-mutant and 8 wild-type mice) from seven different litters. When all data were combined, P values were not significant (fig. 6A). We show the actual data, to allow comparisons among littermates. The range of variation (“biological noise”) is quite striking. Although selective presentation of a subset of these data could yield statistically significant results, by studying a larger number of animals, we were unable to confirm that expression of Dlx5 and Dlx6 is increased in Mecp2-mutant brain.
Figure 6. .
No increase in Dlx5 and Dlx6 expression in Mecp2-mutant mouse brain. A, Expression of Dlx5 and Dlx6 genes in frontal cortex of Mecp2 mutants (mut) compared with that of wild-type (wt) males from seven different litters, at age 7 wk (litters 1–5) or 9 wk (litters 6 and 7). For litters with more than one wild type or more than one mutant, individual animals are identified by different symbols (designated wt1 and wt2 or mut1 and mut2, respectively). Real-time qRT-PCR data from each mouse were converted to relative fold changes by comparison with a normalized sample, a wild-type mouse whose level of gene expression was closest to the mean value for all wild-type data—that is, wt1 in litter 3 for Dlx5 and wt1 in litter 6 for Dlx6. P values calculated by two-tailed, unequal variance are P=.4984 for Dlx5 and P=.5134 for Dlx6. P values calculated by one-tailed, unequal variance are P=.2492 for Dlx5 and P=.2567 for Dlx6. The plots illustrate the wide variability within and between litters. B, Expression of the imprinted gene Peg3, studied in the same forebrain samples used in fig. 6A, by qRT-PCR with two different primer sets (Peg3A and Peg3B). Expression levels were normalized to the same housekeeping gene, Rps28. The wide range of expression levels in individual mice and the similarity of patterns obtained with Peg3A and Peg3B primer sets indicate biological noise rather than technical variation.
To assess whether a similar range of variation would be observed for expression of an imprinted gene, we performed qRT-PCR analyses on the same mouse forebrain samples, with two different intron-spanning primer sets for Peg3 (fig. 6B). An increase in Peg3 expression in mutants was seen in some litters but not in others. The similarity of patterns obtained with the Peg3A and Peg3B primer sets suggests that technical variability plays a minor role. The variation has a biological basis and is unrelated to the Mecp2 genotype. Therefore, we saw no justification for engaging in chromatin modification and chromatin loop assays to evaluate the remaining aspects of the model described in the previous report. The data we obtained argue against the foundation of the proposed model. DLX5 is not imprinted, and there is no relaxation of imprinting in RTT cell lines or brains and no consistent overexpression of Dlx5 and Dlx6 in Mecp2-mutant mouse brain. It is highly unlikely, therefore, that DLX5 is a direct target of MeCP2.
Discussion
A previous report identified DLX5 as a target of MeCP2 and drew wide-ranging conclusions linking RTT with genomic imprinting. The purported “loss of imprinting” of DLX5 was speculated to have implications for GABA neurotransmission and for autism.24 Numerous articles have cited the conclusions of Horike et al.24 as facts, with consequences for research on autism, RTT, Angelman syndrome, and genomic imprinting.36–43 It is the purpose of this article to set the record straight. We provide unequivocal evidence against the claim that DLX5 is imprinted in humans and is a direct target of MeCP2 modulation. As summarized in table 3, we report consistent biallelic expression in interspecies SCHs, LCLs, and cortex from normal donors, with no difference between them and RTT samples. We report the first evidence that DLX6 is not imprinted either. This result is expected, since Dlx5 and Dlx6 are coordinately regulated under the control of Dlx1 and Dlx2 via conserved intergenic enhancers.44 We also point out the inconsistencies of the previous work, calling into doubt the claimed subtle preference of maternal allele expression in normal mouse brain and the reported twofold increase of expression of Dlx5 and Dlx6 in Mecp2-null brain. By comparing forebrain samples from 15 mice from seven litters, we found that expression of Dlx5 and Dlx6 was highly variable, with no significant differences between male Mecp2-mutants and normal male littermates. Thus, our serious attempts to replicate the reported data have yielded absolutely no evidence of imprinting of DLX5 or evidence of any influence of MeCP2 deficiency on the expression of DLX5/Dlx5 or DLX6/Dlx6.
Table 3. .
Human DLX5, DLX6, PEG3, and PEG10 Expression[Note]
| Gene and Sample | Expression | No. of Samples |
| DLX5: | ||
| SCHs | Biallelic | 3 mat and 4 pat |
| RTT LCLs | Biallelic | 3 |
| Normal LCLs | Biallelic | 4 |
| Normal fetal brain | Biallelic | 1 |
| Normal adult cortex | Biallelic | 5 |
| DLX6: | ||
| SCHs | Biallelic | 3 mat and 4 pat |
| Fetal brain | Biallelic | 2 |
| PEG3: | ||
| RTT LCLs | Monoallelic | 3 |
| RTT brain | Monoallelic | 3 |
| Male MeCP2 -/Y brain | Monoallelic | 2 |
| PEG10: | ||
| RTT brain | Monoallelic | 2 |
| Male MeCP2 -/Y brain | Monoallelic | 1 |
Note.— mat = maternal; pat = paternal.
Therefore, any postulated models to explain the nonreproducible data, such as “silent chromatin loop formation,”24(p38) are unfounded. Attempting to explain their finding of increased Dlx5 and Dlx6 expression in Mecp2-deficient mouse brain, Horike et al.24 identified a small region in an intron that contains a single methylated CpG site to which MeCP2 binds. Histone-acetylation and histone-methylation patterns associated with this region were reported to be different between mutant and wild-type mouse brains. It was, therefore, claimed that the chromatin modification differences at this site result in increased Dlx5 expression. On the basis of their prior work with the nuclear matrix binding protein SATB1 (i.e., special AT-rich sequence binding 1) that regulates genes by folding chromatin into loop domains in thymocytes,45 Horike et al.24 then used a chromosome conformation capture (3C) method to generate evidence of differential chromatin loop formation in wild-type versus Mecp2-mutant brains. The proposed model of an 11-kb loop of “silent chromatin” in normal cells and its absence in Mecp2-deficient cells does not, however, correlate with the difference in allele-specific Dlx5 expression reported in the same article. If it were true that MeCP2 facilitates a chromatin loop formation that induces transcriptionally silent chromatin, this should lead to reduced expression of Dlx5 and Dlx6 in normal brain compared with in Mecp2-null brain, without any allelic difference, because the CpG in the intron to which MeCP2 presumably binds is equally methylated on both alleles.24 The model proposed is inconsistent with the fact that Dlx5 is expressed at high levels in normal forebrain. If the proposed silencing mechanism were active in a brain region, such as the cerebellum, where Dlx5 is not expressed, then MeCP2 deficiency should lead to Dlx5 expression in the mutant cerebellum. But Horike et al.24 reported only qRT-PCR data for frontal cortex from 8-wk-old Mecp2-mutant mouse. In our cerebellum microarray studies, Dlx5 expression was not increased in 2-wk-old, 4-wk-old, and 8-wk-old Mecp2 mutants,17 and the level of Dlx5 expression in cerebellum was too low to be reliably measured by use of qRT-PCR. Therefore, the proposed mode of MeCP2 modulation of Dlx5 expression does not fit the cerebellum expression data either.
That MeCP2 may function in chromatin compaction was first proposed by Georgel et al.,46 an article not cited by Horike et al.24 By elegant in vitro studies, Georgel et al. showed that MeCP2 assembles novel secondary chromatin structures from nucleosome arrays but does so independent of DNA methylation. They identified the C-terminal domain of MeCP2 as the region necessary for the nucleosome compaction and suggested that the ability of MeCP2 to silence chromatin may be related to its effect on large chromatin organization. Recently, complex interactions of MeCP2 with DNA and chromatin were revealed by the study of mutant constructs, and they could be divided into methylation-dependent and methylation-independent interactions, with multiple chromatin binding sites identified.47 The functional implications of these interactions are yet unknown.
Expression of Human Genes in Rodent × Human SCHs That Retain a Reduced Number of Human Chromosomes
When the inability to reproduce published data is reported, the discussion needs to focus on experimental design and methodology. Imprinting of human DLX5 was first reported by Okita el al.,28 who studied expression in SCHs containing virally tagged single human chromosomes. In contrast, we found biallelic expression of DLX5 in two recently generated sets of SCHs that segregate human chromosomes randomly. Historically, SCHs were a very useful tool in genetic research. The first decade of human gene mapping relied on the expression of human genes located on the human chromosomes retained in interspecies hybrid cell lines. Distinction between the human and rodent gene products was achieved by electrophoretic separation of proteins that either were stained for activity by substrate binding, in cases of enzymes, or were immunologically detected with human-specific antibodies.48 Faithful expression of human genes in the rodent cell environment, regardless of the number or identity of other human chromosomes present, led to the assignment of numerous gene loci to chromosomes or chromosome regions.49 Extensive work by many researchers solidified the notion that the rodent genome contains all the transcriptional machinery required for expression of human genes from individual human chromosomes retained.
The use of rodent × human SCHs to detect allele-specific expression of imprinted genes was pioneered by Meguro et al.50 By use of microcell-mediated chromosome-transfer methods, the authors generated monochromosomal hybrids with mouse A9 cells that retained only a single virally tagged human chromosome 15 of either parental origin. Working with these cell lines, Meguro et al. reported that the three GABAA receptor subunit genes in the Prader-Willi syndrome/Angelman syndrome deletion region (GABRB3, GABRA5, and GABRG3) are imprinted and paternally expressed. This finding, although still widely cited, was not confirmed in a later study that used a large set of preexisting mouse A9 × human SCHs.51 Gabriel et al.51 validated the general approach, however, by demonstrating that imprinted expression patterns were faithfully retained in SCHs containing either maternal or paternal copies of chromosomes 11 and 15, although the allele-specific DNA methylation patterns were rather variable.
In 1999 and 2001, a Japanese group reported that they made large sets of monochromosomal hybrids that would allow the systematic detection of imprinted genes and other epigenetic phenomena.52,53 A subset of these hybrids was used to study the imprinted gene cluster on chromosome 7 and led to the report that DLX5 is imprinted,28 a result we could not confirm. The successful use of SCH lines for the identification of imprinted genes relies on several prerequisites. First, the parental origin of the single human chromosome retained needs to be reliably identified. Second, the transcription unit to be studied has to be retained intact, because hybrid cells, cultured over extended periods, may undergo chromosome losses, rearrangements, and mutations. Third, the method for detection of expression must reliably distinguish the human and the rodent gene products. In the studies reported here, we used well-characterized hybrid cells that were derived from two unrelated human donors for whom the parental origin of chromosome 7 alleles had been unequivocally established by genotyping all four parents. The SCH lines had been in culture for only a limited time. Originally, we chose two SCHs from each donor that had retained the paternal chromosome 7 and two that had retained the maternal copy, but one of the four lines with the maternal chromosome 7 subsequently lost this chromosome, as determined by PCR amplification of gDNA. Our DLX5 primers were specific to cDNA, not amplification of gDNA. Our primers also were designed with sufficient mismatches against the mouse sequence, to ensure that no mouse transcript would be amplified, and we used mouse brain RNA as a negative control. We detected unambiguous amplification of only human-specific transcripts of DLX5 and DLX6 in SCH lines containing either the maternal (three lines) or the paternal (four lines) chromosome 7, contradicting the findings of Okita et al.28 We note that other data in their article are doubtful as well. Although the authors show imprinted paternal-specific expression for PEG10, confirming previous studies in humans and mice, they report biallelic expression for SGCE. This gene is well known to be imprinted and paternally expressed in human and mouse.54–57 Okita et al. further claimed paternal-only expression for paraoxonase 1 (PON1), a gene that was not confirmed to be imprinted in a subsequent study.58
Use of LCLs for Imprinting Studies
LCLs are derived from peripheral blood B cells by transformation with Epstein-Barr virus.59 They are immortal, although variable in growth rates. Given their transformed nature, LCLs may express tissue-specific genes that are not normally expressed in untransformed B cells. Therefore, the study of neurodevelopmental genes, like DLX5 and DLX6, that are expressed ectopically and at a very low level in LCLs, could lead to failure to amplify RT-PCR products from both alleles (as reported by Okita et al.28 and Horike et al.24). Horike et al. reported monoallelic expression of DLX5 in two normal LCLs and biallelic expression in three of four RTT LCLs. In our experience, only after multiple optimization steps were we able to document consistent biallelic expression of this ectopically expressed locus in all normal control and RTT LCLs. This point is very well illustrated in a recent article by Itaba-Matsumoto et al.,39 which reported studies of 12 LCLs from individuals with RTT and identified MECP2 mutations who were heterozygous for DLX5 c.*163dupC/rs5886002. The authors estimated the degree of X-inactivation mosaicism in these LCLs by deducing relative mutant/wild-type MECP2 expression levels from cDNA sequence tracings. Six LCLs were completely skewed and expressed only wild-type MECP2. Three of those showed biallelic DLX5 expression. Moreover, of five LCLs with 25%–75% mutant MECP2 expression, two showed biallelic DLX5 expression, as did some LCLs from unaffected individuals. Itaba-Matsumoto et al. concluded that they could not reproduce the data reported by Horike et al., but only with regard to MECP2 mutation-type–specific loss of imprinting of DLX5. Itaba-Matsumoto et al.39 did not question the imprinting status of this gene in normal cells and wrote in their abstract, “a sample with high expression of mutated MECP2 in TRD mutation showed bialleic [sic] expression of DLX5 suggesting loss of imprinting.”39 Since these authors were able to amplify only one DLX5 allele in 50% of their cell lines, with no relationship to mutant MECP2 expression levels, their data suggest technical difficulties with RT-PCR caused by low DLX5 expression levels in LCLs.
Although lymphoblast DNA may acquire differential methylation patterns after establishment in culture,60 LCLs have been used successfully for global gene-expression analyses.61 Previous studies reported that monoallelic expression patterns of imprinted genes on chromosomes 11 and 15 are maintained in LCLs.51 Moreover, monoallelic expression was reported for imprinted genes in LCLs from individuals with RTT.20 To further validate the use of LCLs for imprinting studies, we analyzed the expression of PEG3, an imprinted locus on chromosome 19. We found PEG3 to be strictly monoallelically expressed in RTT LCLs that were completely skewed for X inactivation, expressing either 100% mutant or 100% wild-type MeCP2.
Partial Imprinting or Allelic Imbalance
In studies of the forebrain, in which DLX5 is highly expressed, low cDNA copy number should not be an issue for transcript amplification. Indeed, biallelic expression was found in two of three human brain samples studied by Okita et al.28 However, on the basis of inspection of band intensities on sequencing gels, the authors concluded that expression is unequal, with preferential expression of a single allele of unknown parental origin. Our studies revealed unambiguous expression of both DLX5 alleles in six cortex samples from heterozygous individuals, ranging in age from 18 fetal wk to 70 years. Likewise, DLX6 was biallelically expressed in two fetal brains that were heterozygous for a known and a novel SNP, respectively. Therefore, DLX5 and DLX6 are not imprinted in human brain, and we have not observed apparent unequal expression.
Consistent with our results in human, Dlx5 was reported to be biallelically expressed in multiple brain regions, with no allele-specific preference, in F1 mice of interspecies crosses.26 By using the same approach, Horike et al., however, reported “preferential expression from the maternal allele”24(p33) in control mouse brains. This pattern was said to be changed to “equal expression” of Dlx5 alleles in a single Mecp2-mutant brain. This result led the authors to conclude that “MeCP2 deficiency leads to complete loss of relaxed imprinting in brain.”24(p33) The evidence consists of restriction-enzyme digestion of RT-PCR products from heterozygous templates. In this system, quantitation of alleles is difficult because, at the end of the PCR cycles, when the sample is cooling down, denaturation and renaturation may occur, leading to the formation of heteroduplex molecules that are not susceptible to restriction-enzyme cleavage. This phenomenon results in the larger uncleaved band appearing relatively stronger. In addition, the cleaved allele is represented by two smaller bands whose combined fluorescent intensity should approximate that of the uncleaved allele. The pattern expected from a template with both alleles equally represented is illustrated by the gDNA results for the four imprinted genes in figure 2b in the work of Horike et al.,24 in which the largest fragment is always the most intense one. In figure 2a, a single Mecp2-mutant RNA sample is shown with exactly the same intensity of two bands, representing the 636-bp uncut and 533-bp cut Dlx5 allelic fragments. The smaller 103-bp restriction fragment, representing part of the cleavable allele, is cut off. This result would not be expected if both alleles were expressed equally. Therefore, the data do not support the claim of parent-of-origin–specific allelic imbalance of Dlx5 expression in normal brain and a role for MeCP2 in maintaining a maternal preference indicating relaxed imprinting.
Allelic variation in gene expression for nonimprinted genes is increasingly being recognized. The magnitude varies from a 1.3-fold to 9-fold difference in 20%–50% of the genes tested.62–66 Interestingly, allelic transcript variants may follow Mendelian inheritance because of variation in cis-acting elements or “regulatory SNPs.”67 Therefore, unequal allelic expression levels of any gene should not be casually interpreted as “partial imprinting.” Confusion could arise for genes that are imprinted in a cell type–specific fashion, if the sample tested were a mixture of cells affected by the imprinting and cells that are not. There is no evidence that the genes discussed here fall into this category.
Horike et al.24 provided circumstantial evidence in support of the fact that Dlx5 is not imprinted in mouse brain, by showing that there are no DMRs in the vicinity of Dlx5/Dlx6 and that the promoters of both genes are unmethylated on both alleles. Evolutionary considerations lend additional support. As epigenetic phenomena evolve in mammalian species, there are well-established epigenetic differences between humans and mice. In humans, ∼15% of X-linked genes are biallelically expressed—that is, escape from X inactivation—which is not the case in mice.68 Likewise, the list of known imprinted genes contains many that are imprinted in mice but biallelically expressed in humans.19
No Role for MeCP2 in Imprinting
To determine whether truly imprinted genes maintain their imprinting status in MeCP2-deficient brain, we studied PEG3 and PEG10 expression in cortex samples from mosaic females with RTT, as well as from nonmosaic males with congenital encephalopathy who are hemizygous for MECP2 mutations. The strictly monoallelic expression of PEG3 and PEG10 confirms that the lack of normal MeCP2 protein does not abolish or relax the imprinting status of imprinted loci (table 3). Ten imprinted transcripts have previously been shown to maintain their imprinting status in MeCP2-deficient tissues from humans and/or mice: H19, IGF2, SNRPN, IPW, and NDN,20 as well as KCNQ1OT1 (or LIT1), Sgce, Peg10, Asb4, and Calcr.24 Although the role of DMRs for the maintenance of imprinting is undisputed, MeCP2 is unlikely to be the CpG binding molecule that translates this methylation signal.
Acknowledgments
We are grateful to Katherine Carlsmith Harrison, Johannes Richter, ChaRandle Jordan, and Anita Beck, for contributing data, and to Alessandra Splendore and Feng Ding, for constructive comments on the manuscript. This work was supported in part by research grants from the National Institutes of Health and the Rett Syndrome Research Foundation (to U.F.) and fellowships from the Deutsche Forschungsgemeinschaft and the Stanford Medical School Chandler Fund (to B.S.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for DLX5 [accession number NM_005221.5], DLX6 [accession number NM_005222.2], and Dlx6 [accession number BC114342.1] reference sequences)
- HGVS Nomenclature for the Description of Sequence Variants, http://www.hgvs.org/mutnomen/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RTT and MECP2)
References
- 1.Rett A (1966) Uber ein eigenartiges hirnatrophisches Syndrom bei Hyperammonamie im Kindesalter. Wien Med Wochenschr 116:723–738 [PubMed] [Google Scholar]
- 2.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23:185–188 10.1038/13810 [DOI] [PubMed] [Google Scholar]
- 3.Francke U (2006) Neurogenetics of MeCP2 deficiency. Nat Clin Pract Neurol 2:212–221 10.1038/ncpneuro0148 [DOI] [PubMed] [Google Scholar]
- 4.Wan M, Lee SS, Zhang X, Houwink-Manville I, Song HR, Amir RE, Budden S, Naidu S, Pereira JL, Lo IF, et al (1999) Rett syndrome and beyond: recurrent spontaneous and familial MECP2 mutations at CpG hotpots. Am J Hum Genet 65:1520–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carney RM, Wolpert CM, Ravan SA, Shahbazian M, Ashley-Koch A, Cuccaro ML, Vance JM, Pericak-Vance MA (2003) Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol 28:205–211 [DOI] [PubMed] [Google Scholar]
- 6.Kankirawatana P, Leonard H, Ellaway C, Scurlock J, Mansour A, Makris CM, Dure LS, Friez M, Lane J, Kiraly-Borri C, et al (2006) Early progressive encephalopathy in boys and MECP2 mutations. Neurology 67:164–166 10.1212/01.wnl.0000223318.28938.45 [DOI] [PubMed] [Google Scholar]
- 7.Giacometti E, Luikenhuis S, Beard C, Jaenisch R (2007) Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proc Natl Acad Sci USA 104:1931–1936 10.1073/pnas.0610593104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guy J, Gan J, Selfridge J, Cobb S, Bird A (2007) Reversal of neurological defects in a mouse model of Rett syndrome. Science 315:1143–1147 10.1126/science.1138389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nan X, Campoy FJ, Bird A (1997) MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88:471–481 10.1016/S0092-8674(00)81887-5 [DOI] [PubMed] [Google Scholar]
- 10.Colantuoni C, Jeon OH, Hyder K, Chenchik A, Khimani AH, Narayanan V, Hoffman EP, Kaufmann WE, Naidu S, Pevsner J (2001) Gene expression profiling in postmortem Rett syndrome brain: differential gene expression and patient classification. Neurobiol Dis 8:847–865 10.1006/nbdi.2001.0428 [DOI] [PubMed] [Google Scholar]
- 11.Tudor M, Akbarian S, Chen RZ, Jaenisch R (2002) Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci USA 99:15536–15541 10.1073/pnas.242566899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Traynor J, Agarwal P, Lazzeroni L, Francke U (2002) Gene expression patterns vary in clonal cell cultures from Rett syndrome females with eight different MECP2 mutations. BMC Med Genet 3:12 10.1186/1471-2350-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ballestar E, Ropero S, Alaminos M, Armstrong J, Setien F, Agrelo R, Fraga MF, Herranz M, Avila S, Pineda M, et al (2005) The impact of MECP2 mutations in the expression patterns of Rett syndrome patients. Hum Genet 116:91–104 10.1007/s00439-004-1200-0 [DOI] [PubMed] [Google Scholar]
- 14.Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, Schulz R, Lipkowitz B, Ropers HH, Holmes MC, et al (2005) Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum Mol Genet 14:2247–2256 10.1093/hmg/ddi229 [DOI] [PubMed] [Google Scholar]
- 15.Delgado IJ, Kim DS, Thatcher KN, LaSalle JM, Van den Veyver IB (2006) Expression profiling of clonal lymphocyte cell cultures from Rett syndrome patients. BMC Med Genet 7:61 10.1186/1471-2350-7-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng V, Matagne V, Banine F, Frerking M, Ohliger P, Budden S, Pevsner J, Dissen GA, Sherman LS, Ojeda SR (2007) FXYD1 is a MeCP2 target gene overexpressed in the brains of Rett syndrome patients and Mecp2-null mice. Hum Mol Genet 16:640–650 10.1093/hmg/ddm007 [DOI] [PubMed] [Google Scholar]
- 17.Jordan C, Li H-H, Kwan H, Francke U (2007) Cerebellar gene expression profiles of mouse models for Rett syndrome reveal novel MeCP2 targets. BMC Med Genet 8:36 10.1186/1471-2350-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reik W, Walter J (1998) Imprinting mechanisms in mammals. Curr Opin Genet Dev 8:154–164 10.1016/S0959-437X(98)80136-6 [DOI] [PubMed] [Google Scholar]
- 19.Morison IM, Ramsay JP, Spencer HG (2005) A census of mammalian imprinting. Trends Genet 21:457–465 10.1016/j.tig.2005.06.008 [DOI] [PubMed] [Google Scholar]
- 20.Balmer D, Arredondo J, Samaco RC, LaSalle JM (2002) MECP2 mutations in Rett syndrome adversely affect lymphocyte growth, but do not affect imprinted gene expression in blood or brain. Hum Genet 110:545–552 10.1007/s00439-002-0724-4 [DOI] [PubMed] [Google Scholar]
- 21.Samaco RC, Hogart A, LaSalle JM (2005) Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum Mol Genet 14:483–492 10.1093/hmg/ddi045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makedonski K, Abuhatzira L, Kaufman Y, Razin A, Shemer R (2005) MeCP2 deficiency in Rett syndrome causes epigenetic aberrations at the PWS/AS imprinting center that affects UBE3A expression. Hum Mol Genet 14:1049–1058 10.1093/hmg/ddi097 [DOI] [PubMed] [Google Scholar]
- 23.Jordan C, Francke U (2006) Ube3a expression is not altered in Mecp2 mutant mice. Hum Mol Genet 15:2210–2215 10.1093/hmg/ddl146 [DOI] [PubMed] [Google Scholar]
- 24.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T (2005) Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet 37:31–40 10.1038/ng1570 [DOI] [PubMed] [Google Scholar]
- 25.Guy J, Hendrich B, Holmes M, Martin JE, Bird A (2001) A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet 27:322–326 10.1038/85899 [DOI] [PubMed] [Google Scholar]
- 26.Kimura MI, Kazuki Y, Kashiwagi A, Kai Y, Abe S, Barbieri O, Levi G, Oshimura M (2004) Dlx5, the mouse homologue of the human-imprinted DLX5 gene, is biallelically expressed in the mouse brain. J Hum Genet 49:273–277 10.1007/s10038-004-0139-2 [DOI] [PubMed] [Google Scholar]
- 27.Stuhmer T, Anderson SA, Ekker M, Rubenstein JL (2002) Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development 129:245–252 [DOI] [PubMed] [Google Scholar]
- 28.Okita C, Meguro M, Hoshiya H, Haruta M, Sakamoto YK, Oshimura M (2003) A new imprinted cluster on the human chromosome 7q21-q31, identified by human-mouse monochromosomal hybrids. Genomics 81:556–559 10.1016/S0888-7543(03)00052-1 [DOI] [PubMed] [Google Scholar]
- 29.Panganiban G, Rubenstein JL (2002) Developmental functions of the Distal-less/Dlx homeobox genes. Development 129:4371–4386 [DOI] [PubMed] [Google Scholar]
- 30.Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G (1999) Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development 126:3795–3809 [DOI] [PubMed] [Google Scholar]
- 31.Depew MJ, Liu JK, Long JE, Presley R, Meneses JJ, Pedersen RA, Rubenstein JL (1999) Dlx5 regulates regional development of the branchial arches and sensory capsules. Development 126:3831–3846 [DOI] [PubMed] [Google Scholar]
- 32.Perera M, Merlo GR, Verardo S, Paleari L, Corte G, Levi G (2004) Defective neuronogenesis in the absence of Dlx5. Mol Cell Neurosci 25:153–161 10.1016/j.mcn.2003.10.004 [DOI] [PubMed] [Google Scholar]
- 33.Yan H, Papadopoulos N, Marra G, Perrera C, Jiricny J, Boland CR, Lynch HT, Chadwick RB, de la Chapelle A, Berg K, et al (2000) Conversion of diploidy to haploidy. Nature 403:723–724 10.1038/35001659 [DOI] [PubMed] [Google Scholar]
- 34.Gallagher RC, Pils B, Albalwi M, Francke U (2002) Evidence for the role of PWCR1/HBII-85 C/D box small nucleolar RNAs in Prader-Willi syndrome. Am J Hum Genet 71:669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.den Dunnen JT, Antonarakis SE (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15:7–12 [DOI] [PubMed] [Google Scholar]
- 36.Hamilton SP, Woo JM, Carlson EJ, Ghanem N, Ekker M, Rubenstein JL (2005) Analysis of four DLX homeobox genes in autistic probands. BMC Genet 6:52 10.1186/1471-2156-6-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schanen NC (2006) Epigenetics of autism spectrum disorders. Hum Mol Genet Spec 2 15:R138–R150 10.1093/hmg/ddl213 [DOI] [PubMed] [Google Scholar]
- 38.Badcock C, Crespi B (2006) Imbalanced genomic imprinting in brain development: an evolutionary basis for the aetiology of autism. J Evol Biol 19:1007–1032 10.1111/j.1420-9101.2006.01091.x [DOI] [PubMed] [Google Scholar]
- 39.Itaba-Matsumoto N, Maegawa S, Yamagata H, Kondo I, Oshimura M, Nanba E (2007) Imprinting status of paternally imprinted DLX5 gene in Japanese patients with Rett syndrome. Brain Dev (electronically published March 17, 2007) (accessed May 19, 2007) [DOI] [PubMed] [Google Scholar]
- 40.Chadwick LH, Wade PA (2007) MeCP2 in Rett syndrome: transcriptional repressor or chromatin architectural protein? Curr Opin Genet Dev 17:121–125 10.1016/j.gde.2007.02.003 [DOI] [PubMed] [Google Scholar]
- 41.Itoh M, Ide S, Takashima S, Kudo S, Nomura Y, Segawa M, Kubota T, Mori H, Tanaka S, Horie H, et al (2007) Methyl CpG-binding protein 2 (a mutation of which causes Rett syndrome) directly regulates insulin-like growth factor binding protein 3 in mouse and human brains. J Neuropathol Exp Neurol 66:117–123 [DOI] [PubMed] [Google Scholar]
- 42.Kuwajima T, Nishimura I, Yoshikawa K (2006) Necdin promotes GABAergic neuron differentiation in cooperation with Dlx homeodomain proteins. J Neurosci 26:5383–5392 10.1523/JNEUROSCI.1262-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bienvenu T, Chelly J (2006) Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat Rev Genet 7:415–426 10.1038/nrg1878 [DOI] [PubMed] [Google Scholar]
- 44.Zhou QP, Le TN, Qiu X, Spencer V, de Melo J, Du G, Plews M, Fonseca M, Sun JM, Davie JR, et al (2004) Identification of a direct Dlx homeodomain target in the developing mouse forebrain and retina by optimization of chromatin immunoprecipitation. Nucleic Acids Res 32:884–892 10.1093/nar/gkh233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T (2002) SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419:641–645 10.1038/nature01084 [DOI] [PubMed] [Google Scholar]
- 46.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC (2003) Chromatin compaction by human MeCP2: assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem 278:32181–32188 10.1074/jbc.M305308200 [DOI] [PubMed] [Google Scholar]
- 47.Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL (2007) Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol Cell Biol 27:864–877 10.1128/MCB.01593-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Francke U, Pellegrino MA (1977) Assignment of the major histocompatibility complex to a region of the short arm of human chromosome 6. Proc Natl Acad Sci USA 74:1147–1151 10.1073/pnas.74.3.1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Francke U, Busby N, Shaw D, Hansen S, Brown MG (1976) Intrachromosomal gene mapping in man: assignment of nucleoside phosphorylase to region 14cen leads to 14q21 by interspecific hybridization of cells with a t(X;14)(p22;q21) translocation. Somatic Cell Genet 2:27–40 10.1007/BF01539240 [DOI] [PubMed] [Google Scholar]
- 50.Meguro M, Mitsuya K, Sui H, Shigenami K, Kugoh H, Nakao M, Oshimura M (1997) Evidence for uniparental, paternal expression of the human GABAA receptor subunit genes, using microcell-mediated chromosome transfer. Hum Mol Genet 6:2127–2133 10.1093/hmg/6.12.2127 [DOI] [PubMed] [Google Scholar]
- 51.Gabriel JM, Higgins MJ, Gebuhr TC, Shows TB, Saitoh S, Nicholls RD (1998) A model system to study genomic imprinting of human genes. Proc Natl Acad Sci USA 95:14857–14862 10.1073/pnas.95.25.14857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kugoh H, Mitsuya K, Meguro M, Shigenami K, Schulz TC, Oshimura M (1999) Mouse A9 cells containing single human chromosomes for analysis of genomic imprinting. DNA Res 6:165–172 10.1093/dnares/6.3.165 [DOI] [PubMed] [Google Scholar]
- 53.Inoue J, Mitsuya K, Maegawa S, Kugoh H, Kadota M, Okamura D, Shinohara T, Nishihara S, Takehara S, Yamauchi K, et al (2001) Construction of 700 human/mouse A9 monochromosomal hybrids and analysis of imprinted genes on human chromosome 6. J Hum Genet 46:137–145 10.1007/s100380170101 [DOI] [PubMed] [Google Scholar]
- 54.Piras G, El Kharroubi A, Kozlov S, Escalante-Alcalde D, Hernandez L, Copeland NG, Gilbert DJ, Jenkins NA, Stewart CL (2000) Zac1 (Lot1), a potential tumor suppressor gene, and the gene for ε-sarcoglycan are maternally imprinted genes: identification by a subtractive screen of novel uniparental fibroblast lines. Mol Cell Biol 20:3308–3315 10.1128/MCB.20.9.3308-3315.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ono R, Kobayashi S, Wagatsuma H, Aisaka K, Kohda T, Kaneko-Ishino T, Ishino F (2001) A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73:232–237 10.1006/geno.2001.6494 [DOI] [PubMed] [Google Scholar]
- 56.Müller B, Hedrich K, Kock N, Dragasevic N, Svetel M, Garrels J, Landt O, Nitschke M, Pramstaller PP, Reik W, et al (2002) Evidence that paternal expression of the ε-sarcoglycan gene accounts for reduced penetrance in myoclonus-dystonia. Am J Hum Genet 71:1303–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grabowski M, Zimprich A, Lorenz-Depiereux B, Kalscheuer V, Asmus F, Gasser T, Meitinger T, Strom TM (2003) The epsilon-sarcoglycan gene (SGCE), mutated in myoclonus-dystonia syndrome, is maternally imprinted. Eur J Hum Genet 11:138–144 10.1038/sj.ejhg.5200938 [DOI] [PubMed] [Google Scholar]
- 58.Ono R, Shiura H, Aburatani H, Kohda T, Kaneko-Ishino T, Ishino F (2003) Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6. Genome Res 13:1696–1705 10.1101/gr.906803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neitzel H (1986) A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet 73:320–326 10.1007/BF00279094 [DOI] [PubMed] [Google Scholar]
- 60.Hannula K, Lipsanen-Nyman M, Scherer SW, Holmberg C, Hoglund P, Kere J (2001) Maternal and paternal chromosomes 7 show differential methylation of many genes in lymphoblast DNA. Genomics 73:1–9 10.1006/geno.2001.6502 [DOI] [PubMed] [Google Scholar]
- 61.McRae AF, Matigian NA, Vadlamudi L, Mulley JC, Mowry B, Martin NG, Berkovic SF, Hayward NK, Visscher PM (2007) Replicated effects of sex and genotype on gene expression in human lymphoblastoid cell lines. Hum Mol Genet 16:364–373 10.1093/hmg/ddl456 [DOI] [PubMed] [Google Scholar]
- 62.Cowles CR, Hirschhorn JN, Altshuler D, Lander ES (2002) Detection of regulatory variation in mouse genes. Nat Genet 32:432–437 10.1038/ng992 [DOI] [PubMed] [Google Scholar]
- 63.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW (2002) Allelic variation in human gene expression. Science 297:1143 10.1126/science.1072545 [DOI] [PubMed] [Google Scholar]
- 64.Lo HS, Wang Z, Hu Y, Yang HH, Gere S, Buetow KH, Lee MP (2003) Allelic variation in gene expression is common in the human genome. Genome Res 13:1855–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bray NJ, Buckland PR, Owen MJ, O’Donovan MC (2003) Cis-acting variation in the expression of a high proportion of genes in human brain. Hum Genet 113:149–153 [DOI] [PubMed] [Google Scholar]
- 66.Pastinen T, Sladek R, Gurd S, Sammak A, Ge B, Lepage P, Lavergne K, Villeneuve A, Gaudin T, Brandstrom H, et al (2004) A survey of genetic and epigenetic variation affecting human gene expression. Physiol Genomics 16:184–193 [DOI] [PubMed] [Google Scholar]
- 67.Doss S, Schadt EE, Drake TA, Lusis AJ (2005) Cis-acting expression quantitative trait loci in mice. Genome Res 15:681–691 10.1101/gr.3216905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heard E, Disteche CM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20:1848–1867 10.1101/gad.1422906 [DOI] [PubMed] [Google Scholar]