Abstract
Large DNA viruses, including giant mimivirus with a 1.2-Mb genome, exhibit numerous orphan genes possessing no database homologs or genes with homologs solely in close members of the same viral family. Due to their solitary nature, the functions and evolutionary origins of those genes remain obscure. We examined sequence features and evolutionary rates of viral family-specific genes in three nucleo-cytoplasmic large DNA virus (NCLDV) lineages. First, we showed that the proportion of family-specific genes does not correlate with sequence divergence rate. Second, position-dependent nucleotide statistics were similar between family-specific genes and the remaining genes in the genome. Third, we showed that the synonymous-to-nonsynonymous substitution ratios in those viruses are at levels comparable to those estimated for vertebrate proteomes. Thus, the vast majority of family-specific genes do not exhibit an accelerated evolutionary rate, and are thus likely to specify functional polypeptides. On the other hand, these family-specific proteins exhibit several distinct properties: (1) they are shorter, (2) they include a larger fraction of predicted transmembrane proteins, and (3) they are enriched in low-complexity sequences. These results suggest that family-specific genes do not correspond to recent horizontal gene transfer. We propose that their characteristic features are the consequences of the specific evolutionary forces shaping the viral gene repertoires in the context of their parasitic lifestyles.
Following the historical achievement in sequencing the 230-kb genome of human cytomegalovirus as early as 1990 (Chee et al. 1990), a number of large DNA viral genomes have been determined (see http://www.giantvirus.org/). Each of those viral genomes encodes hundreds of predicted protein-coding genes, which approach or even exceed in number those of some unicellular genomes. This undoubtedly points to the complexity of molecular mechanisms underlying their parasitic life cycles, at variance with those of small viruses only exhibiting a handful of genes (e.g., RNA viruses or DNA papillomaviruses). However, the molecular basis of the lifecycles of large DNA viruses is mostly uncharacterized due to a large number of putative genes with no predicted functions in their genomes. About half of the predicted genes in the 1.2-Mb Acanthamoeba polyphaga mimivirus genome are ORFans (coined by Fischer and Eisenberg 1999) exhibiting no detectable homologs in the databases (Raoult et al. 2004). Some of those ORFans might originate in over-predictions of genes. However, a recent proteomics study identified 45 ORFan-derived proteins in its viral particle and suggested a minor contribution of gene prediction errors to the large number of ORFans in mimivirus (Renesto et al. 2006).
The existence of numerous unique genes in large DNA viruses is puzzling, given that many of the remaining genes exhibit readily identifiable sequence similarities spanning entire sequences in distantly related organisms (e.g., DNA polymerases, ribonucleotide reductases, etc.). According to a classical view of viral evolution, the abundance of unique genes in a virus or a group of viruses can be seen as a consequence of fast sequence evolution due to a high mutation rate and relaxed functional constraints (McGeoch and Cook 1994; Sakaoka et al. 1994; McGeoch et al. 2000; Hughes 2002). However, there have been no studies appraising this scenario for diverse large DNA viral genomes. In addition, this “fast-evolution” view somewhat conflicts with another classical view that sees viruses as “gene pickpockets” (Moreira and Lopez-Garcia 2005), frequently stealing genes from their hosts; if their genomes are mostly made of laterally transferred host genes, then homology should be detected for most of the encoded genes (Ogata et al. 2005). An ad hoc combination of these two factors is required to explain the property of the current viral gene repertoires. More recent studies (Daubin et al. 2003; Daubin and Ochman 2004; Forterre 2006) suggested that viruses are rather sources (than recipients or vehicles) of genes for cellular organisms (in contrast, see Yin and Fischer 2006). Thus, a large viral genome may act as an invention factory for new genes. With an increasing body of available sequence data of large DNA viruses, it now becomes possible to scrutinize different aspects of the gene repertoires of large DNA viruses to understand their functions and origins.
Large DNA viruses with a genome exceeding 100 kb have been identified in different families of eukaryotic viruses (herpesviruses, baculoviruses, ascoviruses, nimaviruses, and five others described below) as well as in tailed viruses infecting bacteria (myoviruses and siphoviruses). Genomics has been potent in clarifying evolution patterns within respective viral families (Herniou et al. 2001; McLysaght et al. 2003), though establishing the evolutionary relationships between viral families is a greater challenge due to the scarcity of homologous sequences conserved across viral families (Shackelton and Holmes 2004). Recent structural studies begin to reveal possible common ancestries between several eukaryotic and prokaryotic virus lineages (Benson et al. 2004; Baker et al. 2005; Khayat et al. 2005).
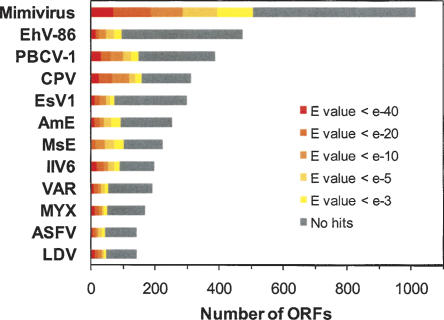
Five eukaryotic viral families (poxviruses, phycodnaviruses, iridoviruses, asfarviruses, and mimiviruses), however, share a relatively large number of conserved core genes, pointing to their ancestral relationships (Iyer et al. 2001, 2006). Furthermore, the genomes of mimivirus and phycodnaviruses are the largest ones among the sequenced viral genomes. Thus, these viruses are of particular interest to study evolutionary patterns of giant DNA viral genomes. These viruses are collectively called as nucleo-cytoplasmic large DNA viruses (NCLDVs). NCLDVs infect a wide range of hosts. They parasitize freshwater amoebae, diverse unicellular and multi-cellular algae, insects, fishes, amphibians, birds, and mammals. NCLDVs exhibit different morphological features in their viral particles. Their genomes vary 10-fold in size (100 kbp to 1.2 Mbp) and exhibit different topologies (i.e., linear versus circular). Some NCLDVs are known to integrate their genome into the host genome but others are not. Thus one can expect a large molecular diversity in their parasitic life cycles. However, the nature of the diversity is largely unknown. Several authors consider that the origin of NCLDVs is relatively recent among other DNA viruses (Koonin 2005; Claverie 2006), though they probably arose before the divergence of major eukaryotic kingdoms (Villarreal and DeFilippis 2000; Raoult et al. 2004). The genetic richness of NCLDVs is revealed by the large proportion (50%–80%) of the predicted genes specific to respective viral families (BLAST E-value < 10−3; Fig. 1). Those genes are either ORFans or sequences exhibiting similarity only in the members of the same viral family. On the other hand, most (75%–100%) of the remaining genes exhibit recognizable similarities to genes from cellular organisms.
Figure 1.
BLAST similarity search results for the predicted proteomes of NCLDVs. Species abbreviations are as follows: LDV, Lymphocystis disease virus; ASFV, African swine fever virus; MYX, Myxoma virus; VAR, Variola virus; IIV6, Invertebrate iridescent virus 6; MsE, Melanoplus sanguinipes entomopoxvirus; AmE, Amsacta moorei entomopoxvirus; EsV1, Ectocarpus siliculosus virus 1; CPV, Canarypox virus; PBCV-1, Paramecium bursaria Chlorella virus 1; EhV-86, Emiliania huxleyi virus 86; Mimivirus, Acanthamoeba polyphaga mimivirus.
In this study, we investigate sequence features and evolutionary rates of viral family-specific genes using three NCLDV lineages: Paramecium bursaria chlorella virus 1 (PBCV-1; the Phycodnaviridae family; database accession no. NC_000852), Myxoma virus (MYX; the Poxviridae family; NC_001132), and variola virus (VAR; the Poxviridae family; NC_001611). For each of these three viruses, closely related genome sequences are available: Paramecium bursaria chlorella virus NY2A (NY2A; DQ491002), Rabbit fibroma virus (Shope fibroma virus, SFV; NC_001266), and Vaccinia virus (VV; NC_006998), respectively. The availability of these close genomes allowed the estimation of evolutionary parameters. Our study demonstrates that the family-specific genes do not differ from other genes in terms of evolutionary rates and nucleotide compositions. However, they exhibit marked differences in size, the predicted capacity to encode transmembrane proteins, and low-complexity sequences.
Results
Lack of correlation between the identification of homologs and the rate of sequence divergence
Some protein sequences evolve faster than others. Fast-evolving protein sequences can rapidly enter the twilight zone of similarity assessment and thus contribute to the abundance of unique genes in NCLDVs. Thus, we first investigated the dependence of similarity detection upon the rate of protein sequence divergence.
We prepared sets of putative orthologs for three viral lineages (PBCV-1/NY2A, MYX/SFV, and VAR/VV) and computed nonsynonymous substitution rate, Ka, for each ortholog pair. Reliable estimation of Ka requires a moderate level of sequence difference between protein coding genes under comparison. Our analysis on the evolutionary rates was restricted to 192 ortholog pairs for PBCV-1/NY2A, 122 for MYX/SFV, and 76 for VAR/VV (see Methods). The average nucleotide sequence identities of the orthologs were 79%, 86%, and 96% for PBCV-1/NY2A, MYX/SFV, and VAR/VV lineages, respectively. We also performed BLAST (Altschul et al. 1997) searches for the open reading frames (ORFs) of PBCV-1, MYX, and VAR against the UniProt protein sequence database (Wu et al. 2006), and defined viral family-specific ORFs, that is, those exhibiting no significant database hit (E-value < 10−3) outside the viral family of the query virus. Other ORFs exhibited significant database hits to proteins from cellular organisms or distantly related viruses. We denote the former as “NORFs” (with a prefix “N” for a Narrow taxonomic distribution) and the latter as “WORFs” (with a prefix “W” for a Wide taxonomic distribution). If sequence divergence rate is a major factor governing the detection of sequence similarity in distant organisms, we should observe fewer NORFs for the class of proteins with lower Ka values. We acknowledge that, due to a relatively high level of sequence divergence between PBCV-1 and NY2A, conclusions derived from the PBCV-1/NY2A comparison should be corroborated with the data from the MYX/SFV and VAR/VV comparisons.
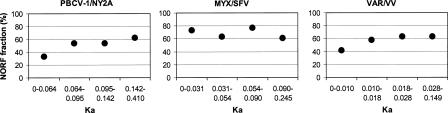
We divided each of the ortholog sets into four classes according to the computed Ka values, so that each class contains one-fourth of the orthologs. We also computed the proportions of NORFs in each ortholog class. Contrary to the expectation, we found no coherent association between the NORF proportions and Ka values. For two viral lineages (MYX/SFV and VAR/VV), we observed no statistically significant correlation between NORF proportions and Ka ranges. For PBCV-1/NY2A orthologs, the NORF proportions were statistically significantly dependent on the Ka ranges (Fisher’s exact test, P < 0.05; Fig. 2). However, the dependence was feeble; when we removed the first Ka category (i.e., most slowly evolving genes), the dependence became no more significant. It is also notable that the numbers of NORFs in the most slowly evolving classes are not negligible (16 for PBCV-1/NY2A, 22 for MYX/SFV, eight for VAR/VV) for all of the three viral lineages. These results suggest that the protein sequence divergence rate estimated by closely related species has little explanatory power for the abundance of family-specific genes in these NCLDVs.
Figure 2.
Lack of correlation between the proportions of family-specific genes and Ka.
Selective strength: Family-specific proteins (NORFs) versus proteins with remote homologs (WORFs)
The heterogeneity in the sequence divergence rate within a proteome is mainly due to variable mutation rates across different genomic loci and variable strength of selection acting on proteins possessing various biochemical/biophysical properties (Li 1997; Rocha 2006). The nonsynonymous-to-synonymous substitution ratio (i.e., ω = Ka/Ks, termed as the “acceptance rate”) is a widely employed estimator of selective strength at the protein level (Miyata and Yasunaga 1980; Nei and Gojobori 1986; Yang et al. 2000), where Ks is thought to correlate with the nonsynonymous “mutation” rate (but see Akashi 1995). Because amino acid replacements in a protein can be detrimental for an organism, ORFs specifying functional polypeptides tend to exhibit ω below one as a result of counter selection against a fraction of nonsynonymous mutations. In contrast, ORFs under a weak selection or those lacking functions (e.g., pseudogenes) tend to exhibit ω values close to one, and ORFs under diversifying positive selection can occasionally exhibit ω values above one.
For all the examined orthologs, the ω values were below one, with most orthologs with ω statistically significantly smaller than one (likelihood ratio test, P < 0.01; Table 1). We found no significant differences in Ka, Ks, and ω between the two ORF classes, except that NORFs showed a larger average Ka than WORFs in the PBCV-1/NY2A comparison (Table 2). More importantly, NORFs exhibited Ka and Ks ranges that largely overlap with those of WORFs, revealing numerous NORFs evolving at the rate (in both synonymous and nonsynonymous sites) comparable to many WORFs (Supplemental Fig. S1).
Table 1.
Number of orthologs exhibiting ω not significantly differing from one
Proportions to the total number of examined orthologs are indicated in parentheses.
Table 2.
Ka, Ks, and ω values for three NCLDV lineages
aThe number of analyzed orthologs.
bThe distributions of Ka were significantly different between WORFs and NORFs (U-test; P < 0.01).
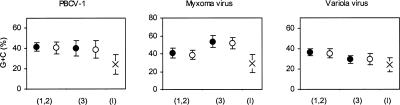
We compared G+C content between NORFs and WORFs with the use of all the ORFs identified in the PBCV-1, MYX, and VAR genomes. G+C content was slightly different between the two ORF classes; NORFs were 1%∼2% less G+C rich than WORFs (P < 0.05; Fig. 3). However, NORFs were markedly different in G+C composition compared with intergenic sequences (9%∼16% differences; P < 0.01). It is notable that the variation in G+C composition across different codon positions is very similar between NORFs and WORFs (Fig. 3). The similar nucleotide composition patterns and sequence divergence rates between the two classes of ORF suggest that the vast majority of NORFs specify bona fide functional proteins evolving under similar levels of purifying selection as WORFs.
Figure 3.
Average G+C compositions of WORFs (filled circles), NORFs (open circles), and intergenic sequences (crosses). For WORFs and NORFs, the G+C compositions at the first and second positions (1,2) and the third positions (3) are separately computed. Bars correspond to a standard deviation. For intergenic sequences, those ≥20 nt were analyzed.
Selective strength: Viral proteomes versus vertebrate proteomes
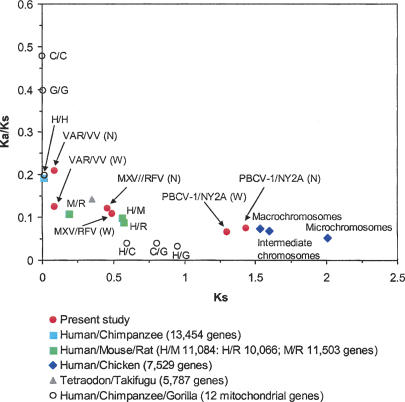
Do NCLDVs differ from cellular organisms in terms of the strength of selection acting on their proteomes? To address this issue, we gathered published genome-scale ω values for several vertebrate genome pairs (Hasegawa et al. 1998; International Chicken Genome Sequencing Consortium 2004; Jaillon et al. 2004; Rat Genome Sequencing Project Consortium 2004; The Chimpanzee Sequencing and Analysis Consortium 2005) and compared them with those estimated for the three viral lineages. Recently, it has become recognized that ω values tend to be larger for closely related genomes than distantly related ones due to a lag in the removal of slightly deleterious mutations from the population (Ho et al. 2005; Penny 2005; Rocha et al. 2006). To take this effect into account, we plotted medians of ω against medians of Ks (Fig. 4). As expected, ω increased with decreasing Ks. After recognizing this dependence of ω on Ks, however, there was no marked difference in ω between the NCLDVs and vertebrates. Thus on the average, the viral proteomes are under comparable levels of selection pressure as the vertebrate proteomes.
Figure 4.
Proteome-scale Ka/Ks for the three NCLDV lineages and several vertebrate pairs with complete genomes. For the NCLDVs, (N) and (W) denote those for NORFs and WORFs, respectively.
As selective strength varies across proteins depending on their functions, it is of interest to compare ω between viruses and vertebrates using genes encoding similar functions. We took genes for B-type DNA polymerases, ribonucleotide reductase small and large subunits, TFII-like transcription factors, mRNA capping enzymes, dUTPases from the three NCLDV lineages, and their homologs from human, mouse, and rat (Fig. 5). There was no recognizable systematic difference in ω between the viral genes and their cellular homologs. However, the ω values vary across proteins as well as among different organism pairs. A notable example is the case for the B-type DNA polymerases. The ω values of the poxvirus DNA polymerases (MYX/SFV and VAR/VV) are three times larger than the ω value of the phycodnavirus lineage (PBCV-1/NY2A). Interestingly, eukaryotic DNA polymerase α catalytic subunits (replication initiation) exhibit larger ω values than δ catalytic subunits (replication elongation). According to a published phylogenetic tree (Villarreal and DeFilippis 2000), poxvirus DNA polymerases are more closely related to eukaryotic α catalytic subunits than to δ, while phycodnavirus DNA polymerases are located at the root of eukaryotic δ subunits. Thus, in this case, the variation in ω may mainly be due to different functional requirements for the subgroups of the homologs (i.e., α-like and δ-like DNA polymerases).
Figure 5.
Comparisons of Ka/Ks values between NCLDV genes and vertebrate homologs.
Overall the traditional view that viral proteomes evolve under more relaxed constraints than cellular organisms does not hold for the three analyzed NCLDV lineages and is probably not true also for other NCLDVs.
Characteristics of NORFs
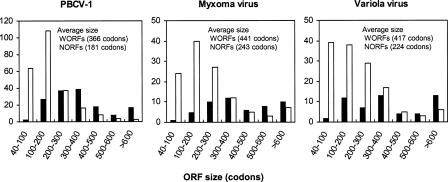
We compared several sequence features between NORFs and WORFs with the use of all the ORFs identified in the PBCV-1, MYX, and VAR genomes. The most prominent feature that was different between the two ORF classes was their sizes. For all the three NCLDV viruses, NORFs were significantly smaller than WORFs (P < 0.01; Fig. 6). On the average, NORFs were 45%∼50% smaller than WORFs. Over-predictions of genes may contribute to the difference, though the above analyses on evolutionary rates and nucleotide compositions suggested minor proportions of over gene predictions. If we consider ORFs longer than 140 codons, over-predicted genes theoretically represent <1% (see Methods). For those ORFs, NORFs were still significantly smaller (28%∼35%) than WORFs (P < 0.01).
Figure 6.
Size distributions of NORFs (white) and WORFs (black) for PBCV-1, MYX, and VAR.
Low-complexity (i.e., repetitive) sequences were more abundant in the protein sequences derived from NORFs than those from WORFs (Supplemental Fig. S2). For PBCV-1, the average proportion of low-complexity sequences for WORF-derived proteins was 4%, while it was 10% for NORF-derived sequences. The difference was statistically significant (P < 0.01). NORFs of MYX and VAR exhibited the same tendency, albeit statistically nonsignificant in these cases.
NORFs were more likely to encode predicted transmembrane proteins than WORFs (Supplemental Fig. S3). In the case of PBCV-1, 21% of the NORFs were predicted to encode transmembrane proteins, while the proportion was 10% for the WORFs. This enrichment in predicted transmembrane proteins is statistically significant for PBCV-1 and MYX (Fisher’s exact test; P < 0.05), but nonsignificant for VAR. The enrichment of predicted transmembrane proteins in the NORF-derived sequences appears a characteristic independent of the small NORF sizes, as the predicted transmembrane proteins do not significantly differ in size from other predicted proteins (data not shown).
The proportions of predicted secondary structures showed no virtual differences between the NORF- and WORF-encoded proteins (Supplemental Fig. S4). Though not salient, however, the helical property of the NORF-encoded proteins was slightly larger than the WORF-derived proteins (P < 0.05 for MYX and VAR). The composition of the predicted extended structures showed a reversed trend (P < 0.05 for MYX). This tendency remained even if we removed the predicted transmembrane proteins and low-complexity sequence segments from the data set (data not shown). Amino acid compositions were also similar between proteins from NORFs and WORFs (Supplemental Fig. S5). Consistently, we could observe no significant difference in predicted isoelectric points between the proteins from NORFs and those from WORFs (data not shown). These results reinforce our conviction that most NORFs encode real proteins.
Thus, in PBCV-1, MYX, and VAR, NORFs are shorter and likely to encode more predicted transmembrane proteins and low-complexity sequences than WORFs. We confirmed the same trend in all the other nine NCLDV genomes listed in Figure 1, except that no enrichment of predicted transmembrane proteins in the NORF class was observed in two viruses (Lymphocystis disease virus and Ectocarpus siliculosus virus 1).
Size does not totally account for the lack of detectable homologs for the NORFs
The NORFs that we defined by BLAST were markedly shorter than the WORFs on the average. Thus, some of the NORFs may have homologs in the current databases at evolutionary distances comparable with the distances from the WORFs to their database homologs. Such homologs might have been missed by BLAST due to its methodological limits. BLAST E-value is related to alignment score by an exponential function (Karlin and Altschul 1990; Altschul et al. 1997). Alignment score has a tendency to decrease with the size of query sequence. Therefore the significance of BLAST E-value dramatically decreases for smaller query sequences at a given evolutionary distance (measured by a substitution matrix).
We performed a simulation to assess this size effect on our data set, by simultaneously taking into account the abundance of low-complexity sequences in proteins derived from the NORFs. The simulation estimates the proportion of NORFs whose homologs are unrecognizable (due to the size effect plus the abundance of low-complexity sequences) even under the assumption that, for all of the NORFs, homologs are present in the database. This corresponds to the maximal proportion of the NORFs possibly having “hidden” homologs in the current database. On the other hand, the size effect and the low-complexity sequence composition will not be able to explain the lack of detectable homologs for the residual proportion of the NORFs.
First, we prepared a list of the sizes of PBCV-1 NORFs (<400 codons) and a list of positions of the low-complexity sequences in the proteins derived from these NORFs. Then, we randomly extracted parts of protein sequences derived from the PBCV-1 WORFs in a way that their size distribution and low-complexity sequence compositions are exactly the same as those for the NORFs (see Methods for details). These artificial amino acid sequences were searched against the database using BLAST. On the average, homologs were unrecognizable by BLAST for 41% of the artificial sequences outside Phycodnaviridae (Fig. 7). These failures of homology detection are primarily attributable to their size and low-complexity sequence content. Thus, at the maximum, 41% of the PBCV-1 NORFs may have “hidden” homologs in the current database. In fact, when we used a profile method for the 240 PBCV-1 NORFs, 15 (6%) exhibited significant database hits (i.e., RPS-BLAST searches against the NCBI/CDD database [Wheeler et al. 2006]; E-value < 10−3). The FUGUE program (Shi et al. 2001), a method for recognizing homology between sequences and known protein structures, could reveal nine (4%) additional hits (Z-score > 6) between NORFs and structural profiles. We obtained similar results for MYX (42% failure) and VAR (39%) through the same kind of simulations (Fig. 7). Thus homologous sequences could be present in the current databases but remain undetected for up to 40% of the NORFs due to the difficulty in assessing the statistical significance of short BLAST alignments.
Figure 7.
Homology search results for the simulated amino acid sequences that mimic the size distribution and low-complexity proportions of NORFs. Bars correspond to standard deviations obtained after 10 times simulations.
On the other hand, the lack of detectable remote homologs for the remaining NORFs (>60%) probably originates in other factors than their sizes and low-complexity sequence contents (see Discussion). For instance, the analyzed genomes exhibit a considerable amount of long NORFs (Fig. 6), for which the average sequence characteristics do not appear to account for the lack of detectable homologs. One of the PBCV-1 ORFs (A505L) corresponds to a 484-aa protein sequence. This sequence exhibits no predicted transmembrane region, only 3% of low-complexity sequence content, and regular evolutionary rates (ω = 0.08, Ka = 0.10 for the comparison with NY2A).
Discussion
The vast majority of the NORFs in the three NCLDV lineages most likely specify functional polypeptides as they exhibit regular sequence divergence rates (i.e., Ka/Ks < 1). The lack of detectable remote homologs of the NORFs is thus attributable to poor sampling in the current databases, to the limits in sequence similarity detection methods, and to their bona fide narrow taxonomic distributions.
Given the huge diversity of viral sequences revealed by metagenomics studies (Angly et al. 2006; Yooseph et al. 2007), inadequate sampling of viral genes in the current databases is likely to be a significant factor leading to the high fraction of unique genes in the NCLDV genomes. NORFs may also have homologs in cellular host genomes that are not sequenced. However, this does not apply to poxviruses, whose multicellular eukaryotic hosts (or close relatives) are well represented in the current databases. For protist-infecting NCLDVs, such a possibility cannot be ruled out, although their proteomes do not exhibit a noticeable enrichment in sequences similar to those of their host (Claverie et al. 2006).
Methodological limits in sequence homology recognition arise from different factors. These include sequence divergence rate, gene size, sequence complexity, and the presence/absence of evolutionary stable sequence motifs. Through computer simulations, we showed that at most 40% of the absence of detectable NORF homologs by BLAST could be due to their short sizes combined with the abundance of low-complexity sequences. In contrast, we established that the lack of recognizable homologs for the NCLDV NORFs are not due to especially high sequence divergence rates usually attributed to viruses. In fact, those genes were found to exhibit evolutionary rates similar to those associated with genes with remote homologs (WORFs). This is at odds with bacterial ORFans. Daubin and Ochman found that bacterial ORFans evolve faster and exhibit much lower G+C contents than other genes, and suggested recent exogenous (i.e., viral) origins for bacterial ORFans (Daubin and Ochman 2004). Such an explanation does not hold for the viral NORFs as they exhibit normal nucleotide compositions and standard sequence divergence rates. Therefore, the NORFs might be much older than bacterial ORFans.
Finally, given the large evolutionary distances between virus families (for example, measured on conserved proteins such as DNA polymerases; Villarreal and DeFilippis 2000), some of the NORFs may be truly specific to a virus family, in a sense that their homologs are absent or drastically different in other (cellular or viral) organisms.
Whatever the reasons for the present uniqueness of the NCLDV NORFs, they correspond to the most uncharacterized parts of their genomes, likely to encode virus-specific molecular strategies to take advantages of their hosts. The three NORF sequence features (smallness, repetitiveness, and predicted membrane association) thus might be tied with yet uncharacterized evolutionary forces acting on viral genomes. Repetitiveness could endow particular functional properties to the encoded amino acid sequences as repetitive sequences are often found in nucleic acid-interacting proteins and cytoskeleton components of eukaryotes. Repetitiveness may also be a consequence of the mechanisms (e.g., DNA replication slippage) randomly generating long open reading frames from scratch (Ohno 1987). Shortness could be the result of gene optimization processes, such as the minimization of existing genes. It is also possible that shortness helps encoded proteins to act as functionally specialized compact modules that can be recruited in different contexts (Lupas et al. 2001). Membrane environment may be an important niche for viral proteins as the surface area to volume ratio is small for virus factories compared to cells. Virus factories are special intracellular compartments for genome replication and assembly of viruses, and are surrounded by membranous structures such as the endoplasmic reticulum and mitochondria (Novoa et al. 2005). Viral membrane proteins may function especially at the stages when virus factories have to promote intensive exchanges of metabolites and cofactors through the membranes. Interestingly, recent studies revealed thus far the smallest known functional potassium ion channel encoded in the PBCV-1 genome (Kang et al. 2004), and the smallest functional mitochondrial carrier encoded in the mimivirus genome (Monne et al. 2007).
Genomics has been focusing on cellular life forms, until we recently realized that viruses are probably the most diverse and abundant life form on our planet (Suttle 2005). Most of what we know on genes has been derived from thorough functional studies using a handful of model organisms such as Escherichia coli, yeast, and more recently animal and plant models. Because of their lack of cellular homologs, the functional characterization of viral genes (i.e., NORFs) will now require the same efforts to be specifically directed to viral organisms. These studies may eventually reveal original metabolic pathways, unexpected regulatory mechanisms, and other unanticipated properties of these “virus only” genes.
Methods
Genome data
NCLDV genome sequence data were downloaded from the viral section of the NCBI Reference Sequence (RefSeq) database (Wheeler et al. 2006) as of January 16, 2006, except for the genome sequence of Paramecium bursaria chlorella virus NY2A. NY2A infects the same host Chlorella-NC64A as PBCV-1. Its complete genome sequence (GenBank accession no. DQ491002; 368,683 bp) has been recently determined (Fitzgerald et al. 2007).
ORF identification
We built ORF sets (≥40 codons) for the NCLDV genomes using Glimmer (Delcher et al. 1999). The standard genetic code was used in this search. Genes containing introns, albeit being rare in NCLDVs, were annotated as in the original annotations in RefSeq except for NY2A. Homologous genes in PBCV-1 were used to annotate intron-containing genes of NY2A using GeneWise (Birney et al. 2004) available at the Wise2 server (http://www.ebi.ac.uk/Wise2/). The analyses presented in this study were based on these ORF sets. Total number of the ORFs identified in the genomes of PBCV-1, MYX, and VAR were 389, 170, and 192, respectively. We also performed the same analyses on the original RefSeq annotations, which also generated essentially the same results (data not shown).
We used a simple model to quantify over-predictions of genes. We generated 100 artificial genome sequences for each of PBCV-1, MYX, and VAR, through random shuffling of genomic sequences that keeps their nucleotide frequencies. We counted ORFs (≥40 codons) occurring in these artificial sequences by the EMBOSS/getorf program (Rice et al. 2000). The size distributions of ORFs for the artificial sequences were compared with those identified by Glimmer for the real genomic sequences. This analysis suggests that potential over-predictions of genes will be <1% for ORFs >140 codons for all the three genomes. We consider that this estimate is very conservative due to the simplicity of the model, and that the true over-prediction rate should be much lower even for ORFs <140 codons.
Homology search
The predicted NCLDV protein sequences were searched against the entire UniProt sequence database (Wu et al. 2006) release 7.4 as of April 2006 to identify homologous sequences using BLAST (Altschul et al. 1997) with an E-value threshold of 10−3 and with the default SEG (Wootton 1994) filtering. BLAST matches against sequences from environmental samples were excluded because of the potential uncertainty in their taxonomic origins. Each NCLDV proteome was divided into two classes: WORFs with detectable homologs outside the viral family to which they belong, and NORFs, which are viral family-specific ORFs, possessing no detectable homologs outside the viral family. The proportions of NORFs were 62% (240 ORFs) for PBCV-1, 69% (118) for MYX, and 71% (137) for VAR.
Definition of orthology
An initial list of orthologs for each of three pairs of genomes (PBCV-1/NY2A, MYX/SFV, and VAR/VV) was defined by reciprocal BLAST best hits, with at least 40% amino acid sequence identity and <50% size difference. We refined the ortholog list by retaining only those exhibiting gene order colinearity. For poxviruses (MYX/SFV, VAR/VV), putative orthologs within the terminal inverted repeat regions were excluded.
Estimation of evolutionary rates
The orthologous protein sequences were aligned by MUSCLE (Edgar 2004) and then back-translated into codon alignments. We removed ambiguously aligned regions (due to short repeats or high divergence) by visual inspection of the pairwise sequence alignments. The synonymous (Ks) and nonsynonymous (Ka) substitution rates and their ratio (ω = Ka/Ks) were computed using the maximum likelihood method implemented in the codeml program in the PAML package (Yang 1997), using constant ω for all sites and the codon frequency model F3×4. For some orthologs, these evolutionary parameters could not be estimated due to saturation of nucleotide substitutions or extremely low level of sequence divergence. Those orthologs were discarded from the analyses. Using the remaining orthologs, we prepared three data sets (I, II, and III) based on the percent standard errors (p-SEs) of the estimations of Ka, Ks, and ω. Data set I and II were composed of all the orthologs that exhibited p-SE(Ka), p-SE(Ks), and p-SE(ω) <100% and 50%, respectively. Data set III was derived from the data set II by retaining only the orthologs, of which the codon alignments were ≥150 codons. The analyses of sequence divergence rates were performed for all the three data sets (I, II, and III), which generated essentially the same results. In this manuscript, we only show the results from the data set II (i.e., p-SEs < 50%). To test if an estimated ω significantly differs from one, we employed the likelihood ratio test (using a χ2-distribution with one degree of freedom, P < 0.01) without corrections for multiple tests.
Vertebrate data
The genome-scale Ks and ω data for vertebrate genomes were obtained from published literature. These correspond to the nuclear genome-encoded 13,454 human/chimpanzee orthologs (The Chimpanzee Sequencing and Analysis Consortium 2005), 10,066–11,503 human/mouse/rat orthologs (Rat Genome Sequencing Project Consortium 2004), 7529 human/chicken orthologs (International Chicken Genome Sequencing Consortium 2004), 5787 Tetraodon/Takifugu orthologs (Jaillon et al. 2004), and the mitochondrial genome-encoded 12 human/chimpanzee/gorilla orthologs (Hasegawa et al. 1998). For the nuclear genome data, we used median values. For the mitochondrial data, we used the data obtained from the concatenated alignment of 12 genes. In their original works, the data for the nuclear genome-encoded human/mouse/rat orthologs were computed by the yn00 program in PAML. The data for the Tetraodon/Takifugu orthologs were computed by the PBL method (Li et al. 1985; Pamilo and Bianchi 1993). For other vertebrate genome pairs, Ks and ω were computed by codeml as in our study. All the ω values for individual ortholog pairs were recomputed by codeml. Vertebrate homologs were obtained from the NCBI/HomoloGene database (Wheeler et al. 2006): DNA polymerase δ (HomoloGene ID 2014), DNA polymerase α (6802), ribonucleotide reductase M1 (806), ribonucleotide reductase M2 (20277) and TP53 inducible M2 B (56723), TFIIS (TCEA1: 55984, TCEA2: 68304, TCEA3: 20684), mRNA capping enzyme (37851), dUTPase (31475).
Protein sequence analysis
The SEG program was used to define low-complexity sequences. We used the Phobius program for the prediction of transmembrane proteins (Kall et al. 2004). Protein secondary structures and isoelectric points (pI) were predicted using the garnier and pepstats programs in EMBOSS (Rice et al. 2000). The sequence-structure homology recognition was performed using FUGUE (Shi et al. 2001) against HOMSTRAD (de Bakker et al. 2001) as of July 2006.
Simulation
The following is the procedure to generate a set of partial protein sequences derived from WORFs, that imitate the size distribution and low-complexity sequence compositions of NORF-derived proteins (<400 resides). We first prepared “source” sequences from the WORF-derived protein sequences, by removing low-complexity sequences and by splitting the sequences into pieces (when applicable) at the locations of their low-complexity regions. We also generated a list of the low-complexity regions identified in the proteins derived from NORFs <400 codons. For each of the NORFs with L codons, we selected one sequence (or occasionally two) from the source set, and randomly extracted a partial segment of L-residues from the source sequence. The choice of the source sequence was randomly made from those >2L. When no such source sequence was available due to a long size of the NORF, we concatenated two source sequences randomly chosen so that the sum of their sizes became >2L. This source sequence selection process ensures that the extracted L-residues segment corresponds to no more than 50% of the source sequence. Finally, the regions in the L-residues segment that match to the low-complexity regions identified in the original L-residues protein sequence from the NORF were masked, by replacing amino acid letters with “X,” to imitate the low-complexity sequence property of the NORF. This procedure generates a set of amino acid sequences exhibiting the same size distribution and low-complexity sequence compositions as the NORFs. For each viral species (PBCV-1, MYX, VAR), we generated 10 such sets to obtain the average of the number of BLAST hits against the UniProt database.
Statistical tests
We used the nonparametric Mann-Whitney U statistic to test the differences in the distributions of G+C compositions, protein sequence identities, evolutionary parameters (Ks, Ka, ω), and the compositions of low-complexity sequences and predicted secondary structures. Other statistics that we used in this study were explicitly noted in the manuscript.
Acknowledgments
We thank Prof. Van Etten for giving us an early access to the genome sequence data of NY2A (GenBank DQ491002). We thank Adam Monier for critical reading of the manuscript. This work was partially supported by Marseille-Nice Genopole and the French National Network (RNG).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6358607
References
- Akashi H. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly F.E., Felts B., Breitbart M., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Felts B., Breitbart M., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Breitbart M., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., Chan A.M., Haynes M., Kelley S., Liu H., Haynes M., Kelley S., Liu H., Kelley S., Liu H., Liu H., et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.L., Jiang W., Rixon F.J., Chiu W., Jiang W., Rixon F.J., Chiu W., Rixon F.J., Chiu W., Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson S.D., Bamford J.K., Bamford D.H., Burnett R.M., Bamford J.K., Bamford D.H., Burnett R.M., Bamford D.H., Burnett R.M., Burnett R.M. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R., Clamp M., Durbin R., Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee M.S., Bankier A.T., Beck S., Bohni R., Brown C.M., Cerny R., Horsnell T., Hutchison C.A., Kouzarides T., Martignetti J.A., Bankier A.T., Beck S., Bohni R., Brown C.M., Cerny R., Horsnell T., Hutchison C.A., Kouzarides T., Martignetti J.A., Beck S., Bohni R., Brown C.M., Cerny R., Horsnell T., Hutchison C.A., Kouzarides T., Martignetti J.A., Bohni R., Brown C.M., Cerny R., Horsnell T., Hutchison C.A., Kouzarides T., Martignetti J.A., Brown C.M., Cerny R., Horsnell T., Hutchison C.A., Kouzarides T., Martignetti J.A., Cerny R., Horsnell T., Hutchison C.A., Kouzarides T., Martignetti J.A., Horsnell T., Hutchison C.A., Kouzarides T., Martignetti J.A., Hutchison C.A., Kouzarides T., Martignetti J.A., Kouzarides T., Martignetti J.A., Martignetti J.A., et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Claverie J.M. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J.M., Ogata H., Audic S., Abergel C., Suhre K., Fournier P.E., Ogata H., Audic S., Abergel C., Suhre K., Fournier P.E., Audic S., Abergel C., Suhre K., Fournier P.E., Abergel C., Suhre K., Fournier P.E., Suhre K., Fournier P.E., Fournier P.E. Mimivirus and the emerging concept of “giant” virus. Virus Res. 2006;117:133–144. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Daubin V., Ochman H., Ochman H. Bacterial genomes as new gene homes: The genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V., Lerat E., Perriere G., Lerat E., Perriere G., Perriere G. The source of laterally transferred genes in bacterial genomes. Genome Biol. 2003;4:R57. doi: 10.1186/gb-2003-4-9-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker P.I., Bateman A., Burke D.F., Miguel R.N., Mizuguchi K., Shi J., Shirai H., Blundell T.L., Bateman A., Burke D.F., Miguel R.N., Mizuguchi K., Shi J., Shirai H., Blundell T.L., Burke D.F., Miguel R.N., Mizuguchi K., Shi J., Shirai H., Blundell T.L., Miguel R.N., Mizuguchi K., Shi J., Shirai H., Blundell T.L., Mizuguchi K., Shi J., Shirai H., Blundell T.L., Shi J., Shirai H., Blundell T.L., Shirai H., Blundell T.L., Blundell T.L. HOMSTRAD: Adding sequence information to structure-based alignments of homologous protein families. Bioinformatics. 2001;17:748–749. doi: 10.1093/bioinformatics/17.8.748. [DOI] [PubMed] [Google Scholar]
- Delcher A.L., Harmon D., Kasif S., White O., Salzberg S.L., Harmon D., Kasif S., White O., Salzberg S.L., Kasif S., White O., Salzberg S.L., White O., Salzberg S.L., Salzberg S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D., Eisenberg D., Eisenberg D. Finding families for genomic ORFans. Bioinformatics. 1999;15:759–762. doi: 10.1093/bioinformatics/15.9.759. [DOI] [PubMed] [Google Scholar]
- Fitzgerald L.A., Graves M.V., Li X., Feldblyum T., Nierman W.C., Van Etten J.L., Graves M.V., Li X., Feldblyum T., Nierman W.C., Van Etten J.L., Li X., Feldblyum T., Nierman W.C., Van Etten J.L., Feldblyum T., Nierman W.C., Van Etten J.L., Nierman W.C., Van Etten J.L., Van Etten J.L. Sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A. Virology. 2007;358:472–484. doi: 10.1016/j.virol.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. DNA topoisomerase V: A new fold of mysterious origin. Trends Biotechnol. 2006;24:245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Cao Y., Yang Z., Cao Y., Yang Z., Yang Z. Preponderance of slightly deleterious polymorphism in mitochondrial DNA: Nonsynonymous/synonymous rate ratio is much higher within species than between species. Mol. Biol. Evol. 1998;15:1499–1505. doi: 10.1093/oxfordjournals.molbev.a025877. [DOI] [PubMed] [Google Scholar]
- Herniou E.A., Luque T., Chen X., Vlak J.M., Winstanley D., Cory J.S., O'Reilly D.R., Luque T., Chen X., Vlak J.M., Winstanley D., Cory J.S., O'Reilly D.R., Chen X., Vlak J.M., Winstanley D., Cory J.S., O'Reilly D.R., Vlak J.M., Winstanley D., Cory J.S., O'Reilly D.R., Winstanley D., Cory J.S., O'Reilly D.R., Cory J.S., O'Reilly D.R., O'Reilly D.R. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 2001;75:8117–8126. doi: 10.1128/JVI.75.17.8117-8126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.Y., Phillips M.J., Cooper A., Drummond A.J., Phillips M.J., Cooper A., Drummond A.J., Cooper A., Drummond A.J., Drummond A.J. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- Hughes A.L. Origin and evolution of viral interleukin-10 and other DNA virus genes with vertebrate homologues. J. Mol. Evol. 2002;54:90–101. doi: 10.1007/s00239-001-0021-1. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Iyer L.M., Aravind L., Koonin E.V., Aravind L., Koonin E.V., Koonin E.V. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L.M., Balaji S., Koonin E.V., Aravind L., Balaji S., Koonin E.V., Aravind L., Koonin E.V., Aravind L., Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Jaillon O., Aury J.M., Brunet F., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Aury J.M., Brunet F., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Brunet F., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Fischer C., Ozouf-Costaz C., Bernot A., Ozouf-Costaz C., Bernot A., Bernot A., et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Kall L., Krogh A., Sonnhammer E.L., Krogh A., Sonnhammer E.L., Sonnhammer E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kang M., Moroni A., Gazzarrini S., DiFrancesco D., Thiel G., Severino M., Van Etten J.L., Moroni A., Gazzarrini S., DiFrancesco D., Thiel G., Severino M., Van Etten J.L., Gazzarrini S., DiFrancesco D., Thiel G., Severino M., Van Etten J.L., DiFrancesco D., Thiel G., Severino M., Van Etten J.L., Thiel G., Severino M., Van Etten J.L., Severino M., Van Etten J.L., Van Etten J.L. Small potassium ion channel proteins encoded by chlorella viruses. Proc. Natl. Acad. Sci. 2004;101:5318–5324. doi: 10.1073/pnas.0307824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Altschul S.F., Altschul S.F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayat R., Tang L., Larson E.T., Lawrence C.M., Young M., Johnson J.E., Tang L., Larson E.T., Lawrence C.M., Young M., Johnson J.E., Larson E.T., Lawrence C.M., Young M., Johnson J.E., Lawrence C.M., Young M., Johnson J.E., Young M., Johnson J.E., Johnson J.E. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc. Natl. Acad. Sci. 2005;102:18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. Virology: Gulliver among the Lilliputians. Curr. Biol. 2005;15:R167–R169. doi: 10.1016/j.cub.2005.02.042. [DOI] [PubMed] [Google Scholar]
- Li W.H. Molecular evolution. Sinauer Associates; Sunderland, MA: 1997. [Google Scholar]
- Li W.H., Wu C.I., Luo C.C., Wu C.I., Luo C.C., Luo C.C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Lupas A.N., Ponting C.P., Russell R.B., Ponting C.P., Russell R.B., Russell R.B. On the evolution of protein folds: Are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J. Struct. Biol. 2001;134:191–203. doi: 10.1006/jsbi.2001.4393. [DOI] [PubMed] [Google Scholar]
- McGeoch D.J., Cook S., Cook S. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J. Mol. Biol. 1994;238:9–22. doi: 10.1006/jmbi.1994.1264. [DOI] [PubMed] [Google Scholar]
- McGeoch D.J., Dolan A., Ralph A.C., Dolan A., Ralph A.C., Ralph A.C. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 2000;74:10401–10406. doi: 10.1128/jvi.74.22.10401-10406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A., Baldi P.F., Gaut B.S., Baldi P.F., Gaut B.S., Gaut B.S. Extensive gene gain associated with adaptive evolution of poxviruses. Proc. Natl. Acad. Sci. 2003;100:15655–15660. doi: 10.1073/pnas.2136653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Yasunaga T., Yasunaga T. Molecular evolution of mRNA: A method for estimating evolutionary rates of synonymous and amino acid substitutions from homologous nucleotide sequences and its application. J. Mol. Evol. 1980;16:23–36. doi: 10.1007/BF01732067. [DOI] [PubMed] [Google Scholar]
- Monne M., Robinson A.J., Boes C., Harbour M.E., Fearnley I.M., Kunji E.R., Robinson A.J., Boes C., Harbour M.E., Fearnley I.M., Kunji E.R., Boes C., Harbour M.E., Fearnley I.M., Kunji E.R., Harbour M.E., Fearnley I.M., Kunji E.R., Fearnley I.M., Kunji E.R., Kunji E.R. The mimivirus genome encodes a mitochondrial carrier that transports dATP and dTTP. J. Virol. 2007;81:3181–3186. doi: 10.1128/JVI.02386-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D., Lopez-Garcia P., Lopez-Garcia P. Comment on “The 1.2-megabase genome sequence of Mimivirus.”. Science. 2005;308:1114. doi: 10.1126/science.1111195. [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Novoa R.R., Calderita G., Arranz R., Fontana J., Granzow H., Risco C., Calderita G., Arranz R., Fontana J., Granzow H., Risco C., Arranz R., Fontana J., Granzow H., Risco C., Fontana J., Granzow H., Risco C., Granzow H., Risco C., Risco C. Virus factories: Associations of cell organelles for viral replication and morphogenesis. Biol. Cell. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H., Abergel C., Raoult D., Claverie J.M., Abergel C., Raoult D., Claverie J.M., Raoult D., Claverie J.M., Claverie J.M. Response to comment on “the 1.2-megabase genome sequence of Mimivirus”. Science. 2005;308:1114b. [Google Scholar]
- Ohno S. Evolution from primordial oligomeric repeats to modern coding sequences. J. Mol. Evol. 1987;25:325–329. doi: 10.1007/BF02603117. [DOI] [PubMed] [Google Scholar]
- Pamilo P., Bianchi N.O., Bianchi N.O. Evolution of the Zfx and Zfy genes: Rates and interdependence between the genes. Mol. Biol. Evol. 1993;10:271–281. doi: 10.1093/oxfordjournals.molbev.a040003. [DOI] [PubMed] [Google Scholar]
- Penny D. Evolutionary biology: Relativity for molecular clocks. Nature. 2005;436:183–184. doi: 10.1038/436183a. [DOI] [PubMed] [Google Scholar]
- Raoult D., Audic S., Robert C., Abergel C., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J.M., Audic S., Robert C., Abergel C., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J.M., Robert C., Abergel C., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J.M., Abergel C., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J.M., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J.M., Ogata H., La Scola B., Suzan M., Claverie J.M., La Scola B., Suzan M., Claverie J.M., Suzan M., Claverie J.M., Claverie J.M. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Renesto P., Abergel C., Decloquement P., Moinier D., Azza S., Ogata H., Fourquet P., Gorvel J.P., Claverie J.M., Abergel C., Decloquement P., Moinier D., Azza S., Ogata H., Fourquet P., Gorvel J.P., Claverie J.M., Decloquement P., Moinier D., Azza S., Ogata H., Fourquet P., Gorvel J.P., Claverie J.M., Moinier D., Azza S., Ogata H., Fourquet P., Gorvel J.P., Claverie J.M., Azza S., Ogata H., Fourquet P., Gorvel J.P., Claverie J.M., Ogata H., Fourquet P., Gorvel J.P., Claverie J.M., Fourquet P., Gorvel J.P., Claverie J.M., Gorvel J.P., Claverie J.M., Claverie J.M. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J. Virol. 2006;80:11678–11685. doi: 10.1128/JVI.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A., Longden I., Bleasby A., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rocha E.P. The quest for the universals of protein evolution. Trends Genet. 2006;22:412–416. doi: 10.1016/j.tig.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Rocha E.P., Smith J.M., Hurst L.D., Holden M.T., Cooper J.E., Smith N.H., Feil E.J., Smith J.M., Hurst L.D., Holden M.T., Cooper J.E., Smith N.H., Feil E.J., Hurst L.D., Holden M.T., Cooper J.E., Smith N.H., Feil E.J., Holden M.T., Cooper J.E., Smith N.H., Feil E.J., Cooper J.E., Smith N.H., Feil E.J., Smith N.H., Feil E.J., Feil E.J. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J. Theor. Biol. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Sakaoka H., Kurita K., Iida Y., Takada S., Umene K., Kim Y.T., Ren C.S., Nahmias A.J., Kurita K., Iida Y., Takada S., Umene K., Kim Y.T., Ren C.S., Nahmias A.J., Iida Y., Takada S., Umene K., Kim Y.T., Ren C.S., Nahmias A.J., Takada S., Umene K., Kim Y.T., Ren C.S., Nahmias A.J., Umene K., Kim Y.T., Ren C.S., Nahmias A.J., Kim Y.T., Ren C.S., Nahmias A.J., Ren C.S., Nahmias A.J., Nahmias A.J. Quantitative analysis of genomic polymorphism of herpes simplex virus type 1 strains from six countries: Studies of molecular evolution and molecular epidemiology of the virus. J. Gen. Virol. 1994;75:513–527. doi: 10.1099/0022-1317-75-3-513. [DOI] [PubMed] [Google Scholar]
- Shackelton L.A., Holmes E.C., Holmes E.C. The evolution of large DNA viruses: Combining genomic information of viruses and their hosts. Trends Microbiol. 2004;12:458–465. doi: 10.1016/j.tim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Shi J., Blundell T.L., Mizuguchi K., Blundell T.L., Mizuguchi K., Mizuguchi K. FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- Suttle C.A. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Villarreal L.P., DeFilippis V.R., DeFilippis V.R. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J. Virol. 2000;74:7079–7084. doi: 10.1128/jvi.74.15.7079-7084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Church D.M., DiCuccio M., Edgar R., Federhen S., DiCuccio M., Edgar R., Federhen S., Edgar R., Federhen S., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton J.C. Sequences with 'unusual' amino acid compositions. Curr. Opin. Struct. Biol. 1994;4:413–421. [Google Scholar]
- Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Ferro S., Gasteiger E., Huang H., Lopez R., Gasteiger E., Huang H., Lopez R., Huang H., Lopez R., Lopez R., et al. The Universal Protein Resource (UniProt): An expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., Goldman N., Pedersen A.M., Nielsen R., Goldman N., Pedersen A.M., Goldman N., Pedersen A.M., Pedersen A.M. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Fischer D., Fischer D. On the origin of microbial ORFans: Quantifying the strength of the evidence for viral lateral transfer. BMC Evol. Biol. 2006;6:63. doi: 10.1186/1471-2148-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yooseph S., Sutton G., Rusch D.B., Halpern A.L., Williamson S.J., Remington K., Eisen J.A., Heidelberg K.B., Manning G., Li W., Sutton G., Rusch D.B., Halpern A.L., Williamson S.J., Remington K., Eisen J.A., Heidelberg K.B., Manning G., Li W., Rusch D.B., Halpern A.L., Williamson S.J., Remington K., Eisen J.A., Heidelberg K.B., Manning G., Li W., Halpern A.L., Williamson S.J., Remington K., Eisen J.A., Heidelberg K.B., Manning G., Li W., Williamson S.J., Remington K., Eisen J.A., Heidelberg K.B., Manning G., Li W., Remington K., Eisen J.A., Heidelberg K.B., Manning G., Li W., Eisen J.A., Heidelberg K.B., Manning G., Li W., Heidelberg K.B., Manning G., Li W., Manning G., Li W., Li W., et al. The Sorcerer II Global Ocean Sampling Expedition: Expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]