Figure 2. .
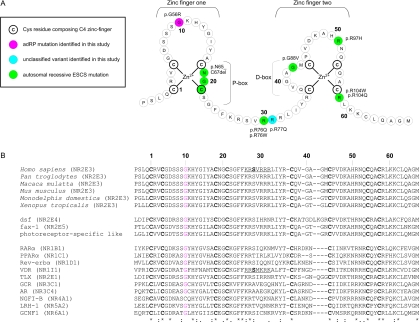
A, Theoretical model of the two C4 zinc fingers located in the DBD of NR2E3. The first zinc finger contains the P-box, in which two or three exposed residues are responsible for half-site sequence recognition. The second zinc finger harbors a dimerization interface for DBDs (D-box) and is involved in recognition of half-site spacing and mutual orientation. A consensus numbering system, starting from the first Cys of the first zinc finger, is used for comparison with other NRs in panel B. B, top rows, Alignment of the human protein sequence of NR2E3 with five orthologs. Middle rows, Three nonhuman NRs belonging to the same subfamily as NR2E3 (dsf, D. melanogaster; fax-1, C. elegans; and photoreceptor-specific like, D. melanogaster). Bottom rows, Human NRs belonging to one of six NR subfamilies: (1) Thyroid hormone receptor–like (RARα, PPARα, Rev-erbα and VDR); (2) HNF4-like (TLX); (3) Estrogen receptor–like (GR and AR); (4) Nerve Growth factor IB receptor–like (NGFI-B); (5) Fushi tarazu-F1–like (LRH-1); and (6) Germ cell nuclear factor–like (GCNF1). The official NR nomenclature is given in parentheses.8 Cys residues composing the C4 zinc-finger motif are indicated in bold. Gly residues in other NRs, corresponding to Gly56 in NR2E3, are indicated in red. Gly56 is conserved in almost all members of the human NR superfamily and in different orthologs, suggesting a functional constraint. Underlined sequences indicate a nuclear localization signal (NLS) in VDR and the corresponding putative NLS in NR2E3. The amino acid Ser, in bold, represents Ser74 and Ser51 in NR2E3 and VDR, respectively (details provided in table 3).