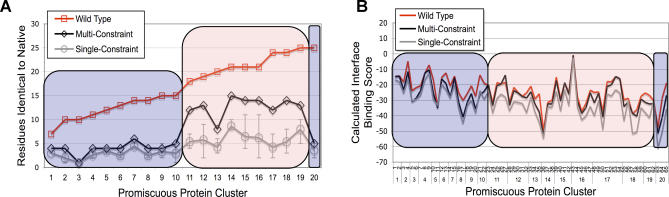
Figure 5. Comparison of Native Amino Acid Recovery and Predicted Binding Scores of Native, Single-Constraint, and Multi-Constraint Sequences.
(A) The number of residues recovered as identical to native are plotted for each promiscuous protein (see Figure 2). For reference, the size of the shared interface is shown for each protein in red. For roughly half the dataset, (group II, pink shading), sequence recovery from the multi-constraint simulations (black) significantly out-performed the average single-constraint recovery (grey). The remaining proteins (group I, blue shading) showed similar native recovery regardless of whether sequences were optimized with respect to one or all characterized partners. Error bars represent the best and worst native sequence recovery in a single-constraint optimization.
(B) Calculated binding scores of native (red), single-constraint (grey), and multi-constraint (black) sequences for each of the 65 complexes examined in this study (see Figure 2). Sequences selected by single- and multi-constraint optimizations often show a favorable decrease in binding score relative to native sequences for group I proteins (blue shading), while multi-constraint binding scores were close to native for group II proteins (pink shading).