Abstract
Lactobacillus delbrueckii mutant Uc-3 utilizes both cellobiose and cellotriose efficiently, converting it into L(+) lactic acid. The enzyme activities of cellobiose and cellotriose utilization were determined for cell extracts, whole cells, and disrupted cells. Aryl-β-glucosidase activity was detected only for whole cells and disrupted cells, suggesting that these activities are cell bound. The mutant produced 90 g/liter of lactic acid from 100 g/liter of cellobiose with 2.25 g/liter/h productivity.
Cellulosic biomass represents an abundant natural renewable carbon resource for the production of valuable fuels and biomaterials for both short- and long-term sustainability. The production of value-added products from such renewable feedstock is a present need and perhaps economically and environmentally feasible process. Lactic acid is a commercially viable product, and world consumption of it is estimated to be more than 60,000 metric tons per year. Lactic acid has a wide range of applications in pharmaceutical, cosmetic, textile, and chemical industries (6, 14, 16, 17). It has the potential to become a commodity chemical as feedstock for biodegradable polymers, oxygenated chemicals, plant growth regulators, environmentally friendly solvents, and special chemical intermediates.
The process for converting cellulosic material into lactic acid is yet not feasible due to the high cost of enzymes involved in cellulose hydrolysis (18, 19, 20) and also to the use of a fastidious organism (10). The process may involve either a two-step process with complete conversion to sugar, followed by fermentation to lactic acid, or a one-step process in which the saccharification of cellulose by cellulases coupled with fermentation to eliminate the inhibition caused by glucose (8, 13).
During the hydrolysis of cellulosic material by cellulases, the main bottlenecks are cellulase inhibition by glucose and cellobiose (strong inhibitors of cellobiohydrolase), which remarkably slow down the rate of hydrolysis. In simultaneous saccharification and fermentation (SSF), glucose inhibition is totally removed but cellobiose inhibition remains as it is (4, 9, 15). The addition of β-glucosidase at the beginning of SSF is recommended for the removal of cellobiose inhibition, but sometimes it is not feasible because of rapid deactivation of enzyme (11). In some cases, cellobiose inhibition was removed by supplementation of the medium with additional cellobiase, leading to a remarkable improvement in lactic acid production in fed-batch SSF (10). However, in simple-batch operations in SSF with cellulase from Trichoderma reesei and Lactobacillus delbrueckii, the supplementation of medium with fresh cellobiase did not improve the overall process (12). To remove these bottlenecks, it is advantageous to use a lactic acid-producing strain that has the ability to utilize both glucose and cellobiose efficiently (9). It is known that some Lactobacillus species utilize cellobiose as a carbon source (5), but very little information is available about lactic acid production from cellobiose. Specifically, to our knowledge, there is no report on the utilization of cellotriose and cello-oligosaccharides by Lactobacillus spp.
In this paper, we describe the efficient utilization of cellobiose and cellotriose by a mutant strain, Lactobacillus delbrueckii Uc-3, for L(+) lactic acid production. The mutant was isolated by UV mutagenesis and selected on the basis of a bigger zone of acid formation on sucrose-based medium (7). We also report the enzyme activities present in Lactobacillus delbrueckii mutant Uc-3 that are involved in cellobiose and cellotriose utilization.
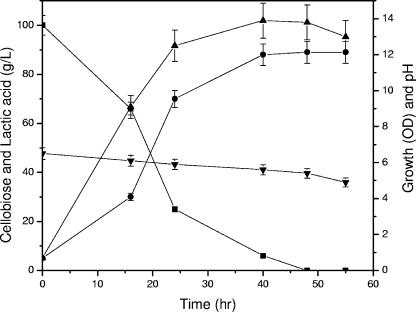
For the evaluation of lactic acid production from cellobiose, experiments were performed in a 250-ml, screw-cap flask at 42°C with shaking at 150 rpm. The flask contained 100 ml production medium consisting of 10 g cellobiose, 4.5 g CaCO3, and 1 g yeast extract. The flasks were inoculated (5% inoculum) with Lactobacillus delbrueckii mutant Uc-3 grown in hydrolyzed, sucrose-based medium (7). After a suitable time interval, the samples for lactic acid and sugar were analyzed by a high-pressure liquid chromatography system equipped with UV or refractive index detectors using an Aminex HPX-87H column (3). The profile of growth (optical density), pH, lactic acid production, and cellobiose utilization is shown in Fig. 1. The maximum amount of lactic acid was produced within 40 h of fermentation, with an increase in optical density from 0.5 to 14 and a decrease in pH from 6.5 to 4.9. The maximum (90 g/liter) amount of lactic acid was produced from 100 g/liter of cellobiose with a 2.25-g/liter/h productivity level and a 0.9-g/g yield. These are the highest productivity and efficiency values reported so far. This strain, therefore, proved to be highly efficient for the conversion of cellobiose to lactic acid and could be exploited at a commercial level. Previously, we reported the production of lactic acid from bagasse-derived cellulose using the same mutant strain. With SSF, we could not detect cellobiose in the fermented broth at any point of time, suggesting the possible presence of β-glucosidase activity in the mutant strain (3).
FIG. 1.
Profile of lactic acid production, growth, pH, and cellobiose utilization during fermentation by Lactobacillus delbrueckii mutant Uc-3 using cellobiose concentration of 100 g/liter. ▪, cellobiose; •, lactic acid; ▴, growth; ▾, pH. Error bars indicate standard deviations. Symbols with error bars that cannot be seen have standard deviations of 5 to 6%.
Separate experiments were carried out to check whether L. delbrueckii Uc-3 utilizes cellotriose or cellooligosaccharides as a sole carbon source. Ten milliliters of medium was prepared in 15-ml, screw-cap tubes containing 1% yeast extract, 25 mg of CaCO3, and 20 mg of cellotriose or cellooligosaccharides separately. The tubes were inoculated with a 5% inoculum grown in 100 g/liter of hydrolyzed sucrose and kept at 42°C under stationary conditions. At 0 h, the samples were analyzed by high-pressure liquid chromatography for the initial concentrations of glucose, fructose, and lactic acid present in the inoculum. Initially, totals of 20 mg glucose, 20 mg fructose, and 10 mg lactic acid were present in the 10 ml of medium. Within 18 h of fermentation, the complete utilization of glucose and fructose led to the production of lactic acid. After 18 h of incubation, the mutant started utilizing cellotriose, and within 30 h of incubation, complete utilization of cellotriose was observed with the production of a proportional amount of lactic acid (Table 1). The cellooligosaccharides were not found to be utilized by this strain. No lactic acid was produced in medium containing cellooligosaccharides, which shows that the mutant does not utilize cellooligosaccharides as a sole carbon source. The total lactic acid produced from 2 g/liter of cellotriose was 1.7 g/liter with a 85% yield. This is the first report which shows that L. delbrueckii used cellotriose as a carbon source and produced lactic acid with a significant yield.
TABLE 1.
Lactic acid production from cellobiose, cellotriose, and cellooligosaccharidesa
| Substrate | Substrate concn (g/liter) | Time of sampling (h) | Lactic acid (g/liter) |
|---|---|---|---|
| Cellobiose | 100 | 40 | 90 ± 4.5 |
| Cellotriose | 2 | 12 | 1.7 ± 0.2 |
| Cellooligosaccharides | 2 | 12 | 0.1 ± 0.12 |
Values are the average of three independent experiments.
We evaluated the capability of this strain to produce lactic acid from cellulose substrates (50 g/liter) like Avicel PH 101, Sigmacell, CP-123, and bagasse-derived cellulose. We carried out SSF experiments using the above-mentioned cellulose substrates by using cellulases derived from a Penicillium janthinellum mutant isolated in our own laboratory (2) and from the L. delbrueckii Uc-3 mutant. The methodology for SSF and the amount of cellulase activities used per gram of substrates were reported earlier (3). The amount of lactic acid produced in SSF from the cellulosic materials varied between 18 and 22 g/liter after 48 h. The level of lactic acid production from bagasse-derived cellulose was 38 g/liter within 60 h of fermentation. We have already reported the complete conversion of this bagasse-derived cellulose to lactic acid with an 80% yield using this strain (3). These results suggest that this Lactobacillus delbrueckii mutant has a potential for the commercial production of lactic acid from biomass material.
This strain was observed to utilize both cellobiose and cellotriose (Table 1). Therefore, we attempted to detect the cellobiose or cellotriose degrading enzymes by using p-nitrophenyl-β-d-glucopyranoside (pNPG), p-nitrophenyl-β-d-cellobioside (pNPC), and p-nitrophenyl-β-d-galactopyranoside (pNPgal) as substrates. The cells were grown in different sugars at 42°C and harvested at late exponential phase by centrifugation. After centrifugation, the supernatant and cells were used for an analysis of aryl-β-glucosidase and aryl-β-galactosidase activities. We could not detect any enzyme activities in the supernatant. The cells were washed three times with citrate phosphate buffer (50 mM, pH 6.0) and suspended in the same buffer. This suspension was used for analyzing cell-bound hydrolytic activities. The assay for aryl-β-glucosidase activity by using pNPG was described earlier (1). The other substrates used were pNPgal and pNPC. One unit of enzyme activity is equivalent to one micromole of p-nitrophenol generated per minute.
Table 2 presents the data of all the enzyme activities in the mutant strain grown in liquid medium with different sugars. All activities were detected in lactose- and cellobiose-grown cells. However, a higher level of aryl-β-glucosidase was observed in cellobiose-grown cells. Other than cellobiose, the lactose-grown cells also exhibited all activities. When cells were grown in glucose and hydrolyzed sucrose-based medium, aryl-β-glucosidase and aryl-β-galactosidase activities were detected in small amounts. No activity on pNPC was detected. These results show that the aryl-β-glucosidase or aryl-β-galactosidase gene could be constitutively expressed, and enhancement in activity was observed when the mutant was grown in either cellobiose or lactose, respectively. The activity on pNPC was detected only when culture was grown in cellobiose or lactose. However, the induction in lactose-grown cells is comparatively less than that in cellobiose-grown cells. This result suggested that aryl-β-glucosidase and aryl β-galactosidase enzymes are independent.
TABLE 2.
Detection of aryl-β-glucosidase activity from Lactobacillus delbrueckii mutant Uc-3 grown in different sugar substratesa
| Enzyme source | Substrate | Enzyme activity (U/g of cells) in:
|
|||
|---|---|---|---|---|---|
| Glucose | Lactose | Hydrolyzed sucrose | Cellobiose | ||
| Cells | pNPG | 2.2 ± 0.20 | 6.5 ± 0.55 | 1.5 ± 0.13 | 11.8 ± 1.0 |
| pNPgal | 4.1 ± 0.35 | 6.0 ± 0.50 | 4.0 ± 0.30 | 3.0 ± 0.25 | |
| pNPC | ND | 1.3 ± 0.12 | ND | 4.5 ± 0.30 | |
| Sonicated cells | pNPG | ND | 4.1 ± 0.35 | ND | 5.5 ± 0.45 |
| pNPgal | 1.0 ± 0.10 | 4.5 ± 0.30 | 1.3 ± 0.10 | 1.7 ± 0.15 | |
| pNPC | ND | 0.8 ± 0.07 | ND | 2.0 ± 0.18 | |
The cells were harvested at the onset of stationary phase for the determination of enzyme activity. Values are the averages of two independent experiments. ND, not detected.
It is noteworthy to say that none of these activities are detected in fermented broth, suggesting the intracellular location of these enzymes. To check the intracellular localization of these enzymes, cells were subjected to sonication (SONICS Vibra cell; model VC 130) in citrate phosphate buffer (pH 6.0, 50 mM) containing 20 mM EDTA, 1 mM dithiothreitol, 10 mM MgCl2, and phenylmethylsulfonyl fluoride (50 μg/ml). The sonication was performed at a 60% amplitude (125 μm) for 5 min by using a 2-mm probe under cold conditions. Almost 90% of the cells were disrupted by this method. The supernatant and sonicated cell debris were analyzed for all above-mentioned enzyme activities. No activities were detected in supernatant, showing that enzymes are not located in cytoplasm. However, all activities (lactose- and cellobiose-grown cells) were detected with sonicated cells (Table 2), suggesting that the enzymes could be cell wall/membrane bound. The activity levels were low compared to those of the intact cells, which could be due to the inactivation of the enzymes during sonication.
In conclusion, the Lactobacillus delbrueckii mutant Uc-3 is a promising strain for the production of lactic acid from cellulosic materials in SSF. Bottlenecks, like feedback inhibition by glucose and cellobiose, were removed by using such a strain, leading to the complete conversion of cellulosic substrates to the products. This strain utilized both cellobiose and cellotriose effectively and produced lactic acid in a homofermentative way. Such an aryl-β-glucosidase, active on both pNPG and pNPC, has not been reported for any of the Lactobacillus species so far. These studies show the potentiality of such a strain, which could produce commodity chemicals from renewable biomass. It could also be possible to generate Lactobacillus strains capable of growing at higher temperatures (50°C), at which the cellulases are most active, which in turn will enhance the efficiency of lactic acid production in SSF.
Acknowledgments
We acknowledge the financial support from the NMITLI Division of the Council of Scientific and Industrial Research, New Delhi, India.
Footnotes
Published ahead of print on 8 June 2007.
REFERENCES
- 1.Adsul, M. G., J. E. Ghule, R. Singh, H. Shaikh, K. B. Bastawde, D. V. Gokhale, and A. J. Varma. 2004. Polysaccharides from bagasse: applications in cellulase and xylanase production. Carbohydr. Polym. 57:67-72. [Google Scholar]
- 2.Adsul, M. G., K. B. Bastawde, A. J. Varma, and D. V. Gokhale. 2007. Strain improvement of Penicillium janthinellum NCIM 1171 for increased cellulase production. Bioresour. Technol. 98:1467-1473. [DOI] [PubMed] [Google Scholar]
- 3.Adsul, M. G., A. J. Varma, and D. V. Gokhale. 2007. Lactic acid production from waste sugarcane bagasse derived cellulose. Green Chem. 9:58-62. [Google Scholar]
- 4.Caminal, G., J. Lopez Santin, and C. Sola. 1985. Kinetic modeling of the enzymatic hydrolysis of pretreated cellulose. Biotechnol. Bioeng. 27:1282-1290. [DOI] [PubMed] [Google Scholar]
- 5.Carr, F. J., D. Chill, and N. Maida. 2002. The lactic acid bacteria: a literature survey. Crit. Rev. Microbiol. 28:281-370. [DOI] [PubMed] [Google Scholar]
- 6.Elezi, O., Y. Kourkoutas, A. A. Koutinas, M. Kanellaki, E. Bezitrzoglou, Y. A. Bernett, and P. Nigam. 2003. Food additive lactic acid production by immobilized cells of Lactobacillus brevis on delignified cellulosic material. J. Agric. Food Chem. 51:5285-5289. [DOI] [PubMed] [Google Scholar]
- 7.Kadam, S. R., S. S. Patil, K. B. Bastawde, J. M. Khire, and D. V. Gokhale. 2006. Strain improvement of Lactobacillus delbrueckii NCIM 2365 for lactic acid production. Process Biochem. 41:120-126. [DOI] [PubMed] [Google Scholar]
- 8.Luo, J., L. Xia, J. Lin, and P. Cen. 1997. Kinetics of simultaneous saccharification and lactic acid fermentation processes. Biotechnol. Prog. 13:762-767. [DOI] [PubMed] [Google Scholar]
- 9.Moldes, A. B., J. L. Alonso, and J. C. Parajo. 2000. Multi-step feeding systems for lactic acid production by simultaneous saccharification and fermentation of processed wood. Bioprocess Eng. 22:175-180. [Google Scholar]
- 10.Moldes, A. B., J. L. Alonso, and J. C. Parajo. 2001. Strategies to improve the bioconversion of processed wood into lactic acid by simultaneous saccharification and fermentation. J. Chem. Technol. Biotechnol. 76:279-284. [Google Scholar]
- 11.Nakasaki, K., and T. Adachi. 2003. Effects of intermittent addition of cellulase for production of l-lactic acid from wastewater sludge by simultaneous saccharification and fermentation. Biotechnol. Bioeng. 82:263-270. [DOI] [PubMed] [Google Scholar]
- 12.Parajo, J. C., J. L. Alonso, and A. B. Moldes. 1997. Production of lactic acid from lignocellulose in a single stage of hydrolysis and fermentation. Food Biotechnol. 11:45-58. [Google Scholar]
- 13.Patel, M. A., M. S. Ou, R. Harbrucker, H. C. Aldrich, M. L. Buszko, L. O. Ingram, and K. T. Shanmugam. 2006. Isolation and characterization of acid-tolerant, thermophilic bacterial biocatalysts for effective fermentation of biomass-derived sugars to lactic acid. Appl. Environ. Microbiol. 72:3228-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Payot, T., Z. Chemaly, and M. Fick. 1999. Lactic acid production by Bacillus coagulans kinetic studies and optimization of culture medium for batch and continuous fermentations. Enzyme Microb. Technol. 24:191-199. [Google Scholar]
- 15.Ramos, L. P., and J. N. Saddler. 1994. Enzyme recycling during fed-batch hydrolysis of cellulose derived from steam-exploded Eucalyptus vitaminalis. Appl. Biochem. Biotechnol. 45:193-207. [Google Scholar]
- 16.Senthuran, A., V. Senthuran, B. Mattiasson, and R. Kaul. 1997. Lactic acid fermentation in a recycle batch reactor using immobilized Lactobacillus casei. Biotechnol. Bioeng. 55:843-853. [DOI] [PubMed] [Google Scholar]
- 17.Venkatesh, K. V. 1997. Simultaneous saccharification and fermentation of cellulose to lactic acid. Bioresour. Technol. 62:91-98. [Google Scholar]
- 18.Wyman, C. E., B. E. Dale, R. T. Elander, M. Holtzapple, M. R. Ladisch, and Y. Y. Lee. 2005. Coordinated development of leading biomass pretreatment technologies. Bioresour. Technol. 96:1959-1966. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, Y. H. P., and L. R. Lynd. 2006. A functionally-based model for hydrolysis of cellulose by fungal cellulase. Biotechnol. Bioeng. 94:888-898. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, Y. H. P., M. Himmel, and J. R. Mielenz. 2006. Outlook for cellulase improvement: screening and selection strategies. Biotechnol. Adv. 22:452-481. [DOI] [PubMed] [Google Scholar]