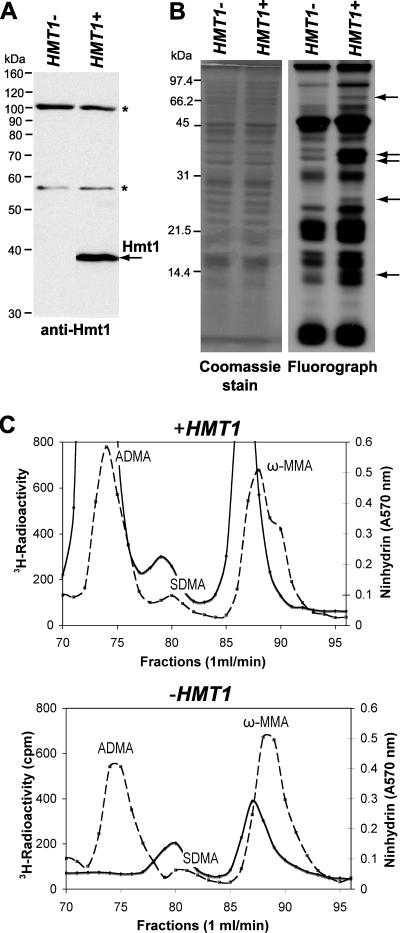
FIG. 2.
C. albicans Hmt1 is not essential and is the major PRMT. (A) hmt1Δ/hmt1Δ (AMC11; HMT1−) and hmt1Δ/hmt1Δ+HMT1 (AMC14; HMT1+) C. albicans strains were created using a PCR-based strategy. Total protein (10 μg) from these strains was analyzed by immunoblotting with anti-ScHmt1 antiserum. (B) C. albicans strains were labeled in vivo with [3H]AdoMet as described in Materials and Methods. Proteins in cell extracts (50 μg) were resolved by SDS gel electrophoresis and analyzed by Coomassie blue staining and fluorography. The dried gel was exposed to Kodak X-Omat AR film at −80°C for 10 days to detect methylated proteins. Arrows indicate major 3H-methylated polypeptides found in the hmt1Δ/hmt1Δ+HMT1 strain (AMC14; HMT1+) expressing the methyltransferase and absent or greatly reduced in the mutant hmt1Δ/hmt1Δ strain (AMC11; HMT1−). (C) Amino acid analysis of C. albicans proteins from lysed cells in vivo labeled with [3H]AdoMet. Acid-hydrolyzed proteins (150 μg) were fractionated on a cation-exchange column as described in Materials and Methods. Radioactivity (3H) (solid line) was determined by scintillation counting. Nonradiolabeled amino acid standards of ADMA, ω-MMA, and SDMA were added to the hydrolysate and detected using a ninhydrin assay (dashed line). The upper panel shows the labeling in cells expressing Hmt1 (AMC14; HMT1+), and the lower panel shows the labeling in cells lacking Hmt1 (AMC11; HMT1−). The radioactivity in the off-scale fractions of the upper panel is 2,625 for fraction 72, 5,707 for fraction 73, and 1,172 for fraction 87. In these experiments, the 3H-labeled methylated derivatives eluted slightly earlier than the nonisotopically labeled standards due to an isotope effect (18).