Abstract
The activation of the ADE regulon genes requires the pair of transcription factors Bas1 and Pho2. In a genome-wide screen for additional regulators of the pathway, strains with mutations in multiple subunits of the chromatin remodeling complexes SAGA and SWI/SNF were uncovered. These mutants exhibited decreased expression of an ADE5,7-lacZ reporter and native ADE compared to the wild-type strains, but the expression of the BAS1 and PHO2 genes was not substantially decreased. An unregulated Bas1-Pho2 fusion protein depended upon SAGA and SWI/SNF activity to promote transcription of a reporter. A significant but low-level association of Gcn5-myc and Snf2-myc with the ADE5,7 promoter was independent of adenine growth conditions and independent of the presence of the activator proteins Bas1 and Pho2. However, the increase in occupancy of Bas1 and Pho2 at ADE5,7 depended on both SAGA and SWI/SNF. The loss of catalytic activity of both SAGA and SWI/SNF complexes in the gcn5Δ snf2Δ double mutant was severely detrimental to ADE-lacZ reporter expression and native ADE gene expression, indicating complementary roles for these complexes. We conclude that Bas1 and Pho2 do not recruit the SAGA and SWI/SNF complexes to the ADE5,7 promoter but that the remodeling complexes are necessary to increase the binding of Bas1 and Pho2 in response to the adenine regulatory signal. Our data support the model that the SAGA and SWI/SNF complexes engage in global surveillance that is necessary for the specific response by Bas1 and Pho2.
The ADE genes of Saccharomyces cerevisiae encode the enzymes of the purine nucleotide biosynthetic pathway responsible for the conversion of 5′-phospho-d-ribosyl-1(β)-pyrophosphate into purine nucleotides. Synthesis of purine nucleotides is metabolically regulated by feedback inhibition of the first step of the purine biosynthetic pathway by the end products of the pathway ATP and ADP (39). At the genetic level, ADE gene expression is transcriptionally repressed when cells are cultured in the presence of purine bases such as adenine (3, 4, 7, 8, 23, 54). Linking the genetic and metabolic regulation are the intermediates of the biosynthetic pathway SAICAR [1-(5′-phosphoribosyl)-4-(N-succinocarboxamide)-5-amino imidazole] and AICAR [1-(5′-phosphoribosyl)-4-carboxamide-5-aminoimidazole]. Cells with a mutation in any step of the pathway prior to SAICAR synthesis exhibit low, nonderepressible expression of ADE genes, whereas cells with a mutation in the steps after SAICAR or AICAR synthesis have high, constitutive ADE gene expression (39). Therefore, the regulatory signals for low levels of purine nucleotides are the pathway intermediates SAICAR and AICAR (39, 40).
Nine ADE genes are up-regulated by the transcription factors Bas1 and Pho2 under derepressing conditions (3, 58, 65). These two factors also regulate the HIS1, HIS4, and HIS7 genes of the histidine biosynthesis pathway and GLN1, SHM2, and MTD1 genes involved in the synthesis of glutamine, glycine, and 10-formyl tetrahydrofolate, respectively (1, 4, 5). Together these 15 genes constitute the ADE regulon, although two recent studies have identified up to 56 genes that Bas1 may bind and regulate (21, 24). Both factors are necessary to stimulate expression, but their DNA binding is differentially regulated. Bas1 binds the ADE upstream activation sequence (UASADE) under both repressing and derepressing conditions (51), but Pho2 binds only under derepressing conditions and this binding depends upon interaction with Bas1 (12, 36, 51). The current model for adenine regulation is that the interaction between Bas1 and Pho2 is promoted by the pathway intermediates SAICAR and AICAR (39, 40, 65); this interaction stabilizes Pho2, with its own activation domain, at the UASADE and unmasks the latent activation domain in Bas1 (12, 36, 65), both of which are necessary to fully activate transcription.
Although Bas1 and Pho2 are required to stimulate ADE regulon expression, how they interact with the transcriptional machinery or chromatin remodeling complexes is not known. At HIS7, Bas1 and Pho2 are thought to promote transcription through Gcn5-dependent histone acetylation (59). PHO5 expression, which is regulated by Pho2 in conjunction with Pho4, is dependent on the Snf2 and Gcn5 subunits of the SWI/SNF and SAGA complexes (27, 52). It has also been shown that Pho2 contacts the chromatin remodeling complex NuA4 and that this interaction is necessary for the expression of PHO5 (29). Thus, several chromatin remodeling complexes may be able to interact with Pho2 and Bas1 in different transcriptional contexts and therefore be relevant as additional regulators of the ADE regulon.
In this study, we follow up on an observation that strains with mutations in several subunits of the SAGA and SWI/SNF chromatin remodeling complexes are sensitive to the adenine analog 4-aminopyrazolo-(3,4d)-pyrimidine (4-APP) (R. J. Rolfes et al., unpublished data). Previous studies of yeast demonstrated that resistance to 4-APP occurs in two different ways. Cells acquired a mutation in the purine-cytosine permease that blocked uptake of the analog (33, 35). Alternatively, cells acquired a specific mutation in the ADE4 gene that produced a feedback-insensitive form of the first enzyme in the purine nucleotide pathway (33-35). These results indicate that a critical balance between de novo synthesis of purine nucleotides and the intracellular concentration of analog-containing nucleotides is necessary for cells to grow on a medium containing 4-APP. Perturbing this balance can lead to 4-APP resistance or sensitivity (Rolfes et al., unpublished). We found that strains that carry mutations in SAGA and SWI/SNF are more sensitive to 4-APP, suggesting a role for these chromatin remodeling complexes in nucleotide metabolism.
SAGA is a multiprotein complex whose Gcn5 subunit encodes a histone acetyltransferase (HAT). Gcn5 acetylates several lysine residues on the histone tails, including K14 on H3 and K8 and K16 on H4 (19). SAGA is also composed of adaptor proteins of the Ada family, TATA binding protein-associated factors, and Spt proteins involved in the structural integrity of the complex (53, 62). The SWI/SNF complex is a highly conserved remodeling complex that utilizes the energy from its ATPase component Snf2 to remodel the local chromatin structure (26, 31, 50).
The recruitment of the HAT complex SAGA has been associated with recruitment of the SWI/SNF complex in many instances (10, 13, 14, 57). In some cases, acetylation by the SAGA component Gcn5 has been shown to increase retention of the SWI/SNF complex on promoters (13). Thus, it was significant that these two complexes were both identified in our screen, as they may carry out their actions in concert at ADE gene promoters. In this report we find that SAGA and SWI/SNF complexes associate with the ADE5,7 promoter, independently of adenine regulation and independently of Bas1 and Pho2. On the other hand, the Gcn5 subunit of SAGA and the Snf2 component of SWI/SNF are required for efficient binding of Bas1 and Pho2 to the UASADE5,7 under derepressing conditions but not under the basal (repressing) conditions. These results demonstrate a role for SAGA and SWI/SNF in increasing the occupancy of the transcriptional activators in response to the adenine regulatory signal.
MATERIALS AND METHODS
Yeast strains.
All strains used in this study are described in Table 1. The homozygous diploid knockout collection was obtained from Research Genetics. To ensure that the strains carried the indicated deletion mutation, each strain selected from the knockout collection for study was analyzed by PCR amplification of the locus (data not shown) using the primers pairs listed on the Saccharomyces Genome Deletion Project website (http://sequence-www.stanford.edu/group/yeast_deletion_project/deletions3.html). We obtained strains FY1352, FY1353, FY1354, and FY1360 from Fred Winston (41) and strains HQY367 and HQY453 from Alan Hinnebusch (38, 64). Strain RR439 is a derivative of FY1360 that was constructed to convert the snf2Δ::LEU2 allele to snf2Δ::leu2::kanMX by transformation with BamHI-restricted plasmid m3926 (61). Transformants were selected for G418 resistance and screened for leucine auxotrophy.
TABLE 1.
Yeast strains
| Name | Genotype | Source or reference |
|---|---|---|
| 4741/4742 | MATa/α his3Δ/his3Δleu2Δ/leu2Δlys2Δ/LYS2 MET15/met15Δura3Δ/ura3Δ | Research Genetics |
| bas1Δ strain | Isogenic to 4741/4742 with bas1Δ::kanMX/bas1Δ::kanMX | Research Genetics |
| gcn5Δ strain | Isogenic to 4741/4742 with gcn5Δ::kanMX/gcn5Δ::kanMX | Research Genetics |
| snf2Δ strain | Isogenic to 4741/4742 with snf2Δ::kanMX/snf2Δ::kanMX | Research Genetics |
| spt3Δ strain | Isogenic to 4741/4742 with spt3Δ::kanMX/spt3Δ::kanMX | Research Genetics |
| spt8Δ strain | Isogenic to 4741/4742 with spt8Δ::kanMX/spt8Δ::kanMX | Research Genetics |
| spt20Δ strain | Isogenic to 4741/4742 with spt20Δ::kanMX/spt20Δ::kanMX | Research Genetics |
| ada1Δ strain | Isogenic to 4741/4742 with ada1Δ::kanMX/ada1Δ::kanMX | Research Genetics |
| ada2Δ strain | Isogenic to 4741/4742 with ada2Δ::kanMX/ada2Δ::kanMX | Research Genetics |
| ada3Δ strain | Isogenic to 4741/4742 with ada3Δ::kanMX/ada3Δ::kanMX | Research Genetics |
| ubp8Δ strain | Isogenic to 4741/4742 with ubp8Δ::kanMX/ubp8Δ::kanMX | Research Genetics |
| sgf73Δ strain | Isogenic to 4741/4742 with sgf73Δ::kanMX/sgf73Δ::kanMX | Research Genetics |
| snf5Δ strain | Isogenic to 4741/4742 with snf5Δ::kanMX/snf5Δ::kanMX | Research Genetics |
| snf11Δ strain | Isogenic to 4741/4742 with snf11Δ::kanMX/snf11Δ::kanMX | Research Genetics |
| taf14Δ strain | Isogenic to 4741/4742 with taf14Δ::kanMX/taf14Δ::kanMX | Research Genetics |
| snf6Δ strain | Isogenic to 4741/4742 with snf6Δ::kanMX/snf6Δ::kanMX | Research Genetics |
| swi3Δ strain | Isogenic to 4741/4742 with swi3Δ::kanMX/swi3Δ::kanMX | Research Genetics |
| FY1352 | MATahis3-Δ200 ura3-52 leu2-Δ1 lys2-173R2 gcn5Δ::HIS3 snf2Δ::LEU2 | 41 |
| FY1353 | MATahis3-Δ200 ura3-52 leu2-Δ1 lys2-173R2 | 41 |
| FY1354 | MATα his3-Δ200 ura3-52 leu2-Δ1 lys2-173R2 gcn5Δ::HIS3 | 41 |
| FY1360 | MATahis3-Δ200 ura3-52 leu2-Δ1 lys2-173R2 snf2Δ::LEU2 | 41 |
| HQY367 | MATa his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 SNF2-myc13::HIS3 | 64 |
| HQY453 | MATa his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 SPT7-myc13::HIS3 | 38 |
| RR376 | MATα BAS1-MYC13::kanMX6 PHO2 ura3-52 | 51 |
| RR409 | MATα BAS1 PHO2-HA3::kanMX6 ura3-52 | 51 |
| RR430 | Isogenic to FY1353 with PHO2-HA3::kanMX | This study |
| RR431 | Isogenic to FY1354 with PHO2-HA3::kanMX | This study |
| RR432 | Isogenic to FY1360 with PHO2-HA3::kanMX | This study |
| RR433 | Isogenic to FY1353 with BAS1-MYC13::kanMX | This study |
| RR434 | Isogenic to FY1354 with BAS1-MYC13::kanMX | This study |
| RR435 | Isogenic to FY1360 with BAS1-MYC13::kanMX | This study |
| RR436 | Isogenic to FY1353 with bas1-10Δ::hisG | This study |
| RR437 | Isogenic to FY1354 with bas1-10Δ::hisG | This study |
| RR439 | Isogenic to FY1360 with snf2Δ::leu2::kanMX | This study |
| RR440 | Isogenic to RR439 with bas1-10Δ::hisG | This study |
| RR441 | Isogenic to HQY367 with bas1-10Δ::hisG | This study |
| RR442 | Isogenic to HQY453 with bas1-10Δ::hisG | This study |
| RR443 | Isogenic to HQY367 with pho2-10Δ::hisG | This study |
| RR444 | Isogenic to HQY453 with pho2-10Δ::hisG | This study |
Strains RR433, RR434, and RR435 are derivatives of FY1353, FY1354, and FY1360, respectively, that carry the BAS1-MYC13 allele, and strains RR430, RR431, and RR432 are derivatives of FY1353, FY1354, and FY1360, respectively, that carry the PHO2-HA3 allele (51). These strains were created by homologous recombination to insert the epitope tag into the chromosomal genes (51). The BAS1-MYC13 module was amplified from genomic DNA of strain RR376 using oligonucleotides JO-205 and JO-206, and the PHO2-HA3 module was amplified from genomic DNA of strain RR409 using oligonucleotides IO-611 and IO-612, as described previously (51). Transformants were selected on yeast extract-peptone-dextrose plates containing 200 μg/ml G418. PCR amplification of the loci from genomic DNA was used to confirm the presence of the tag (data not shown). Expression of the epitope-tagged protein was assessed by immunoblot analysis using anti-myc and antihemagglutinin (anti-HA) antibodies (data not shown).
Strains RR436, RR437, RR440, RR441, and RR442 are derivatives of FY1353, FY1354, RR439, HQY367, and HQY453, respectively, that carry the bas1-10Δ::hisG allele. These strains were generated by replacement of the BAS1 locus with bas1-10Δ::hisG-URA3-hisG using the EcoRI-SphI fragment obtained from pR166 and selection on SC medium lacking uracil (51). The URA3 gene was popped out by culturing the strain on 5-fluoroorotic acid (5-FOA) medium to generate bas1-10Δ::hisG. The presence of the bas1 deletion allele was confirmed by PCR analysis of the genomic locus and by assaying adenine bradytrophy (data not shown).
Strains RR443 and RR444 are derivatives of HQY367 and HQY453, respectively, that carry the pho2-10Δ::hisG allele. These strains were generated by integrative transformation with the EcoRI-SphI fragment obtained from pR167 and selection on SC medium lacking uracil (65). The URA3 gene was popped out by culturing these strains on 5-FOA medium to generate pho2-10Δ::hisG. The presence of the pho2 deletion allele was confirmed by the inability to grow on medium lacking inorganic phosphate and by PCR analysis of the genomic locus (data not shown).
Plasmids.
Plasmid pR109 is a fusion of the lacZ gene to ADE4 at codon 71 and was created by inserting lacZ into the ADE4 gene (65) and subcloning it into a LEU2 CEN6 vector (48) (plasmid construction details available upon request). Plasmid pR115 carries the ADE5,7-lacZ reporter, was constructed by inserting the lacZ gene into the ADE5,7 open reading frame (ORF) and then moving the fusion gene into a LEU2 CEN6 vector, and was described previously (65). Plasmids pR109 and pR115 are similar in that they have large genomic fragment inserts that carry the entire coding region and the entire promoter and regulatory region, extending into the flanking genes. Plasmid pR133, also described previously (65), carries only a small portion (139 bp) of ADE5,7 that carries the regulatory region (UASADE5,7) that was inserted into a heterologous system—the CYC1 core promoter driving the lacZ reporter—developed by L. Guarente and colleagues to characterize UAS elements (11). To distinguish these two types of lacZ reporters, we refer to this latter type as a UASADE5,7-lacZ reporter and the former type as an ADE-lacZ reporter. Plasmids pR166 (51) and pR167 (65) carry the bas1-10Δ and pho2-10Δ alleles, respectively. Plasmid m3926 converts LEU2 to leu2::kanMX through a simple insertion (61). Plasmid pCB286 (1) expresses full-length Bas1, and plasmid B273, obtained from Odd Stokke Gabrielsen, expresses the Bas1-Pho2 fusion protein (36).
Media.
Strains were cultured at 30°C on YPAD solid medium or SD and SC liquid or solid medium containing 2% glucose, 0.17% yeast nitrogen base (Difco), and 0.5% ammonium sulfate supplemented with amino acids as previously described (47). Adenine was added to a final concentration of 0.15 mM where indicated, and 4-APP was added to a final concentration of 75 μM. Medium lacking inorganic phosphate was prepared by precipitation of magnesium phosphate under alkaline conditions, as described previously (45). Medium containing 5-FOA was made by supplementing SC medium with 0.1% 5-FOA. G418 was added to YPAD medium at 200 μg/ml.
β-Galactosidase assays.
β-Galactosidase enzyme assays were performed as previously described (22). Briefly, cells were grown overnight in SC medium containing adenine. Cultures were diluted 1:50 into 50 ml of SC medium with adenine or SC medium lacking adenine and grown for 5 h with rotary agitation at 30°C. Extracts were prepared from pelleted cells by using glass beads, and β-galactosidase activities were measured from clarified whole-cell extracts (22).
4-APP spot test.
Cultures were grown overnight at 30°C overnight in 5 ml of YPAD medium. Cultures were diluted to an optical density at 600 nm (OD600) of 0.2 and allowed to grow for approximately 3 hours. Approximately 1 ml of cells at an OD600 of 0.5 was pelleted and resuspended in 500 μl of sterile water. Serial 10-fold dilutions were prepared, and 5 μl of each dilution was spotted onto the surface of solid medium. Growth was assessed after 48 h of incubation at 30°C.
RNA isolation and Northern blot assays.
Cell cultures were grown overnight in SC medium with adenine. Cultures were diluted to an OD600 of 0.1 in 50 ml and were allowed to grow to an OD600 of 0.5. For derepression, cultures were pelleted, washed twice with SD medium, and resuspended in 50 ml of medium lacking adenine. After 30 min, the cells were pelleted and RNA was isolated using the RNA Isolation Kit (Epicenter); preliminary experiments showed that maximal expression of the ADE5,7 and ADE17 loci occurred at 30 min in the wild-type strain (data not shown). RNA samples were separated on 1.2% formaldehyde gels, blotted to GeneScreen Plus filters, and hybridized to probes (46). The ADE5,7 probe was prepared as described previously (43). Fragments for probes for BAS1, ACT1, and ADE17 detection were derived from PCR amplification of genomic DNA using the following oligonucleotides: BAS1-up (5′-CATGGGCCTCACTGGCTGAG) and BAS1-down (5′-ATCGCTATCCATGTTGTCC), ACT-up (5′-CTTTCTGGAGGAGCAATGATC-3′) and ACT-down (5′-GGATTCCGGTGATGGTGTTAC-3′), and ADE17-up (5′-GAGCAGCTGCTAAGAACCATG-3′) and ADE17-down (5′-ATGGCCAACGCCTCAGGCTC-3′). Radiolabeled probes were prepared using the Random Primed DNA Labeling Kit (Roche) incorporating [α-32P]dATP (43). RNA transcript levels were quantified by phosphorimaging analysis.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as described in reference 9, with the following changes. We added an additional wash of the lysed pellet using fresh FA lysis buffer. DNA in the extracts was fragmented by sonication using a Misonex 3000 sonicator for 30 s on and 30 s off for a total time of 14 min to produce chromatin fragments with an average size of 500 bp, as detected by gel electrophoresis and ethidium bromide staining. Protein G-Dynabeads (Dynal), approximately 2 × 107 in 50 μl, were preincubated with 2 μg of the anti-myc (Santa Cruz) or the anti-HA (Roche) antibody for 3 h at 4°C.
Quantitative, multiplex PCR amplification mixtures contained 1× Platinum Taq polymerase buffer (Invitrogen), 1.5 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate, 1.6 μCi of [α-32P]dATP (Amersham), 0.2 μM POL1 primer pair, 0.2 μM ADE5,7 primer pair, 1.5 U of Platinum Taq polymerase (Invitrogen), and 2 μl of the immunoprecipitated chromatin sample or diluted input DNA (previously determined to be in the linear range for amplification) in a 15-μl reaction mixture volume. PCR parameters were 94°C for 4 min; 94°C for 30 s, 52°C for 30 s, and 65°C for 1 min for 26 cycles; and then 65°C for 5 min. PCR products were resolved on 6% polyacrylamide gels and quantified by phosphorimaging analysis. The sequences of the primer pairs were previously described for POL1 (56) and ADE5,7 (51).
RESULTS
SAGA and SWI/SNF components are important for resistance to adenine analog 4-APP.
In a screen of the yeast deletion collection with the adenine analog 4-APP, we identified ∼50 analog-sensitive mutant strains (Rolfes et al., unpublished). The mutants were divided into functional classes. One class contained genes involved in transcription, including BAS1 and PHO2, which are necessary for expression of the adenylate biosynthetic genes (reviewed in reference 42). This class of mutants also included the genes ADA1, ADA3, GCN5, SPT20, and SPT8, which encode subunits of the SAGA complex, and SNF2, SNF5, and SWI3, which encode subunits of the SWI/SNF complex. Because we identified multiple subunits from both complexes, we hypothesized that these complexes are regulators of ADE gene expression. To confirm and extend this initial observation, we assessed the 4-APP sensitivity of strains carrying mutations in all available identified subunits of the SAGA and SWI/SNF complexes.
As shown in Fig. 1, we found that these eight originally identified strains were sensitive to 4-APP, as indicated by decreased growth of the mutants on SC medium containing 4-APP compared to the growth of the wild-type strain. We also observed sensitivity to 4-APP in five additional strains that carried deletion alleles of the genes SPT3, ADA2, SNF11, TAF14, and SNF6. These 13 mutations encode the majority of the nonessential subunits of the SAGA and SWI/SNF complexes (49, 62).
FIG. 1.
SAGA and SWI/SNF mutant strains are sensitive to the adenine analog 4-APP. Strains with deletion mutations in the SAGA and SWI/SNF complexes were grown to equivalent cell densities, serially diluted (1:10), and spotted on complete medium containing 4-APP, containing adenine, or lacking adenine. Strains were allowed to grow for approximately 48 h and were photographed.
We observed that the sensitivity to 4-APP varied in the different mutant strains. Some of the strains with mutations in the SAGA components exhibited strong sensitivity to 4-APP, including the spt3Δ and spt8Δ mutants, whereas others, like the ada3Δ strain, exhibited only weak sensitivity to 4-APP. Some of the strains had a secondary slow-growth phenotype and an additional 4-APP sensitivity that was very strong for the gcn5Δ mutant and weak for the spt20Δ, ada1Δ, and ada2Δ strains. Strains carrying mutations in the other identified SAGA subunit genes, UBP8, SGF73, and SGF11 (17, 37), were also tested for a 4-APP sensitivity phenotype; however, these strains showed no appreciable sensitivity to 4-APP (Fig. 1).
Likewise, we observed a range of sensitivities to 4-APP in strains with mutations in different components of the SWI/SNF complex. Strong 4-APP sensitivity was detected in the snf2Δ, snf5Δ, and snf6Δ mutant strains, whereas the snf11Δ and taf14Δ mutant strains exhibited only a weak sensitivity to 4-APP. A strain with the swi3Δ mutation was also extremely sensitive to 4-APP, although it was severely growth compromised even on SC medium containing adenine.
Together these results indicated that the absence of catalytic activity found in Gcn5 and Snf2, as well as the loss of key structural subunits provided by Ada1 and Spt20, led to the greatest degree of 4-APP sensitivity. Interestingly, we did not identify subunits from other remodeling complexes such as NuA3, ISWI, or Ino80. These findings indicate that the SAGA and SWI/SNF complexes play a critical role in ADE gene expression.
SAGA and SWI/SNF mutants have decreased ADE gene reporter expression.
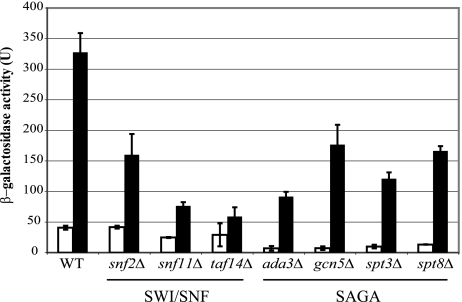
To determine if mutations in SAGA and SWI/SNF affect ADE gene expression, we assayed expression of a lacZ reporter in strains carrying deletion mutations for subunits of the SAGA and SWI/SNF complexes. This lacZ reporter contains binding sites for Bas1 and Pho2 from the ADE5,7 promoter (UASADE5,7) in a heterologous context (CYC1-lacZ). This plasmid was chosen because the insert was extensively characterized by mutational analysis as the minimum necessary for regulation and because it is strongly derepressed by adenine limitation (44). Strains with mutations in subunits of SWI/SNF (snf2Δ, snf11Δ, and taf14Δ) and SAGA (gcn5Δ, spt3Δ, spt8Δ, and ada3Δ) were transformed with this reporter and assayed for effects on expression. We were unable to obtain stable transformants for several of the mutant strains after multiple attempts (swi3Δ, snf5Δ, snf6Δ, ada2Δ, ada1Δ, and spt20Δ strains).
As shown in Fig. 2, we found that each mutant strain was unable to stimulate expression of the UASADE5,7-lacZ reporter to the level of the wild type under derepressing conditions (SC medium lacking adenine). The greatest effect on β-galactosidase activity was observed in the ada3Δ, snf11Δ, spt3Δ, and taf14Δ strains, with expression at approximately 20 to 41% of the wild-type activity. Expression of the reporter was also significantly affected in the snf2Δ, gcn5Δ, and spt8Δ mutant strains, in which β-galactosidase levels decreased to about 56 to 65% of the wild-type level. All of the mutants, with the possible exception of the taf14Δ mutant, responded to adenine limitation by increasing expression of the reporter gene; however, the extent of the derepression in each mutant strain was smaller than that found in the wild type. This observation indicates that the mutant strains were able to generate and receive the intracellular derepression signal to stimulate transcription of the ADE genes but were unable to fully initiate transcription.
FIG. 2.
Expression of a UASADE5,7-lacZ reporter is compromised in SAGA and SWI/SNF mutants. The wild-type strain (BY4741/BY4742) and isogenic strains with mutations in subunits of SWI/SNF (snf2Δ, snf11Δ, and taf14Δ strains) and SAGA (gcn5Δ, spt3Δ, spt8Δ, and ada3Δ strains) were transformed with plasmid pR133 (44), which carries the UASADE5,7-lacZ reporter. Strains were grown in SC medium containing adenine (open bars) or lacking adenine (filled bars). Extracts were prepared from cell pellets, and β-galactosidase activities were measured in duplicate assays on at least three individual isolates. Error bars indicate standard deviations.
Expression of ADE genes is dependent on SAGA and SWI/SNF.
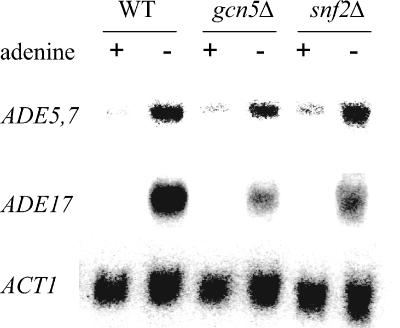
We have shown that SAGA and SWI/SNF components are necessary for efficient lacZ reporter expression driven by the ADE5,7 promoter and regulatory region. To investigate the involvement of SAGA and SWI/SNF in ADE gene expression, we assayed steady-state RNA levels expressed from the native loci in mutant and wild-type strains using Northern analysis. We chose to assay transcript levels from two well-characterized ADE genes, ADE5,7 and ADE17 (3, 39, 44), to represent the ADE regulon and to use the gcn5Δ and snf2Δ mutants defective in the catalytic activities found in each of the complexes. We found that the gcn5Δ and snf2Δ mutant strains were unable to derepress ADE5,7 and ADE17 to the wild-type levels (Fig. 3), and no effect was observed on the repressed levels. In the gcn5Δ mutant, expression of the ADE17 and ADE5,7 loci was reduced to 62 to 63% of the wild-type level, whereas in the snf2Δ mutant, expression was modestly reduced to 71 to 82% of the wild-type level. Thus, the SAGA and SWI/SNF chromatin remodeling complexes are required to achieve full expression of at least these members of the ADE regulon.
FIG. 3.
Expression of ADE5,7 and ADE17 is reduced in gcn5Δ and snf2Δ mutants. RNA was prepared from wild-type (WT) (FY1353) and gcn5Δ (FY1354) and snf2Δ (FY1360) mutant strains that were harvested after 30 min of growth under repressing conditions (SC medium with adenine) and derepressing conditions (SC medium without adenine). RNA was blotted onto nylon membranes and hybridized to probes for ADE5,7, ADE17, and ACT1. Results were quantified by phosphorimaging analysis, and the levels of ADE5,7 and ADE17 were normalized to the reference ACT1 level.
These results show that efficient transcription depends upon the SAGA and SWI/SNF remodeling complexes in addition to the Bas1 and Pho2 transcription factors. Several mechanisms could account for the decrease in the lacZ reporter and native gene expression that we detected in the mutants. The most likely mechanisms are that the Bas1 and Pho2 factors recruit the remodeling complexes that are necessary for the subsequent engagement of RNA polymerase II or that the remodeling complexes are necessary for the binding of Bas1, Pho2, or both to the UASADE. Alternatively, an indirect mechanism might be used if the expression of the activator genes BAS1 and PHO2 or the synthesis of the signaling molecules SAICAR and AICAR depends on the remodeling complexes. We experimentally address these possibilities below.
SAGA and SWI/SNF mutations do not affect BAS1 or PHO2 expression.
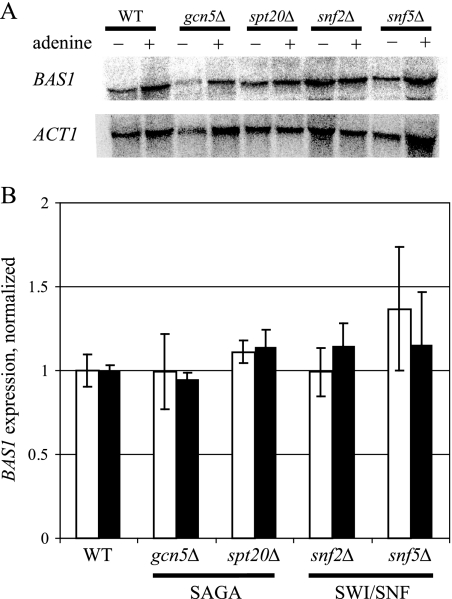
If expression of the BAS1 or PHO2 genes strongly depended on the SAGA and SWI/SNF complexes, then the levels of the activator proteins might be lower in the mutants, decreasing ADE gene expression. To determine if the expression of BAS1 was decreased in SAGA and SWI/SNF mutants, we performed Northern analysis on RNA isolated from strains carrying mutations in the genes encoding structural subunits (spt20Δ and snf5Δ) and catalytic subunits (gcn5Δ and snf2Δ) of both complexes. As shown in Fig. 4, we found that strains carrying the deletion mutations were not significantly affected in the transcription of BAS1. We also observed that the expression of BAS1 was not dependent on exogenous adenine in both the mutant strains and the wild-type strain, consistent with a previous report (65).
FIG. 4.
Expression of BAS1 is not affected in strains with mutations in SAGA and SWI/SNF. (A) The isogenic wild-type (WT) strain and strains carrying mutations in the gcn5 and spt20 genes of SAGA and the snf2 and snf5 genes of SWI/SNF were grown in SC medium containing adenine and SC medium lacking adenine. Total RNA was purified from harvested cells, blotted onto nylon membranes, and hybridized to radiolabeled probes for BAS1 and ACT1. This experiment was performed twice, and results from a representative Northern analysis are shown. (B) The results were quantified by phosphorimaging analysis, and the BAS1/ACT1 ratio was normalized to the wild-type ACT1 level. Samples were analyzed in duplicate, and the error bars indicate the ranges. Open bars, SC medium containing adenine; filled bars, SC medium lacking adenine.
Previous reports demonstrated that PHO2 expression is not affected by mutations in the SPT7 and GCN5 genes of SAGA or in the SNF2 and SWI1 subunits of SWI/SNF (28, 30, 55). We performed Northern analysis on RNA prepared from our strains and observed no difference in expression of PHO2 in gcn5Δ, spt20Δ, snf2Δ, and snf5Δ mutants (data not shown), in agreement with the previously published reports. Together, these results indicate that mutations in catalytic and structural subunits of SAGA and SWI/SNF do not substantially alter the expression of the genes encoding the transcription factors Bas1 and Pho2.
SAGA and SWI/SNF mutants are defective in ADE expression with a constitutively active Bas1-Pho2 fusion protein.
A second explanation for the decrease in ADE gene expression is that the derepression signals SAICAR and AICAR (39) are produced at lower levels in the mutants. Basal expression of the ADE genes is high in Saccharomyces (3, 39); thus, when feedback inhibition is initially relieved, basal levels of the pathway enzymes allow for immediate synthesis of downstream products, including the intracellular signals SAICAR and AICAR. If mutations in subunits of the SAGA and SWI/SNF decreased the basal expression of enzymes prior to formation of SAICAR and AICAR, then the ability to generate the derepression signal might be attenuated, leading to decreased reporter and native gene expression.
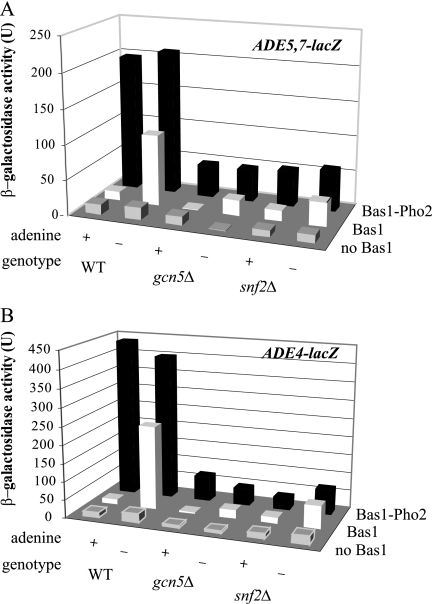
The role of the regulatory signal is to promote interaction between DNA-bound Bas1 and Pho2 (36, 65). This interaction is critical for unmasking latent activation domains in Bas1. To circumvent the need for the signal, we used a Bas1-Pho2 fusion protein (36). This fusion protein constitutively stimulates expression of target genes and is independent of exogenous adenine levels and the intracellular signals (36). To prevent competition between native Bas1 and the Bas1-Pho2 fusion protein for binding sites, we deleted the BAS1 gene in the wild-type and remodeling mutant strains prior to transformation with plasmids that express Bas1 or the Bas1-Pho2 fusion protein and the lacZ reporters. Because the gene encoding the Bas1-Pho2 fusion protein was marked with URA3, we could not use plasmid pR133 as we did above but instead used two different lacZ reporters. The ADE4-lacZ and ADE5,7-lacZ reporters carry the core promoters and regulatory regions from the native ADE4 and ADE5,7 genes, and both reporters are carried on plasmids marked by LEU2. We then asked whether expression of the ADE-lacZ reporter genes was altered in the SAGA and SWI/SNF mutants.
As shown in Fig. 5, in the absence of any Bas1 protein, we found only basal, nonderepressible expression of both the ADE5,7-lacZ and ADE4-lacZ reporters in all strains, whereas strains expressing Bas1 stimulated adenine-regulated expression in the wild-type strain and decreased expression in the remodeling mutants. As expected, the Bas1-Pho2 fusion was not repressible by adenine and it promoted higher reporter gene expression than the Bas1 and Pho2 proteins in the strain with wild-type remodeling complexes (36). Interestingly in the gcn5Δ and snf2Δ mutants, the Bas1-Pho2 fusion protein was able to express the ADE5,7-lacZ reporter to only 20 to 30% of the wild-type level and the ADE4-lacZ reporter to only about 10 to 20% of the wild-type level (Fig. 5A and B). Thus, engineering the transcriptional activators to be independent of the depression signal did not overcome the requirement for the remodeling complexes. These data are consistent with a model in which SAGA and SWI/SNF are involved in promoting transcription after reception of the regulatory signal by the activators, by making the chromatin template more accessible to the transcriptional machinery. However, these data are also consistent with the model that SAGA and SWI/SNF are involved in the binding of the activators to the UASADE, affecting Bas1-Pho2 fusion protein occupancy. Since these roles are not mutually exclusive, the remodelers may have both activities.
FIG. 5.
A constitutively active Bas1-Pho2 fusion protein promotes only low-level expression of ADE-lacZ reporters in gcn5Δ and snf2Δ mutant strains. The wild type (RR436) and strains carrying mutations in gcn5 (RR437) and snf2 (RR439) were transformed with empty vector (YCp50), Bas1 (pR201), or the Bas1-Pho2 fusion (B273), as well as reporter plasmids for ADE5,7-lacZ (pR115 in panel A) or ADE4-lacZ (pR109 in panel B). Strains were grown in SC medium containing or lacking adenine. Extracts were prepared from cell pellets, and β-galactosidase activities were measured in duplicate assays for at least three individual isolates.
SAGA and SWI/SNF are present at the ADE5,7 promoter in both repressing and derepressing conditions.
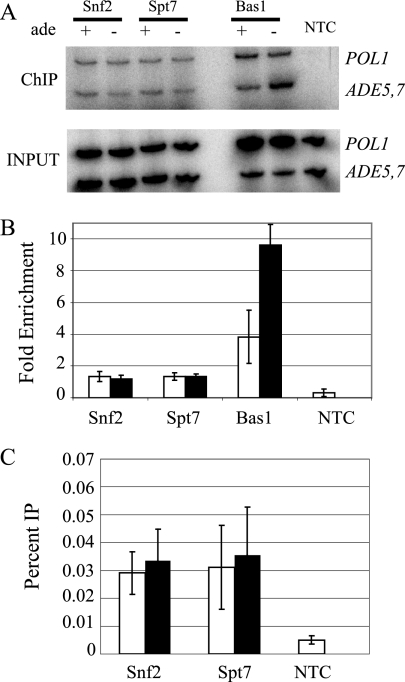
The data presented above suggest that Bas1 and Pho2 are capable of recruiting the SAGA and SWI/SNF complexes. We wanted to determine whether we could detect the presence of these complexes at an ADE gene promoter. We performed ChIP assays, using a tagged subunit found in each remodeling complex, Spt7-myc for SAGA and Snf2-myc for SWI/SNF. These tagged strains were used by other researchers to investigate the association of SAGA and SWI/SNF with the ARG1 and SNZ1 genes (38, 64). We assayed binding of SAGA and SWI/SNF at the promoter of the ADE5,7 gene relative to their binding at the POL1 ORF and asked if their binding increased under the derepressing conditions of SC medium without adenine.
We found that Spt7-myc and Snf2-myc are present at the promoter of the ADE5,7 gene independently of the presence or absence of adenine (Fig. 6A). Relative to the occupancy of Bas1-myc, we found lower association of both Spt7-myc and Snf2-myc, although in both cases we were able to chromatin immunoprecipitate the promoter fragment significantly more highly than in the no-tag control (NTC) (Fig. 6B). We also calculated the percent IP and observed a seven- to ninefold increase in Snf2-myc and Spt7-myc relative to the NTC strains, indicating their presence at the ADE5,7 promoter (Fig. 6C). Somewhat surprisingly, we found that both Spt7-myc and Snf2-myc were able to chromatin immunoprecipitate to POL1, indicating that SAGA and SWI/SNF are almost as equally abundant in the POL1 ORF as they are at ADE5,7 (Fig. 6A). Thus, in comparing the ratio of ChIP at ADE5,7 to that at POL1, we observed a ratio slightly above 1, at about 1.3 for each sample (Fig. 6B), indicating that SAGA and SWI/SNF were only slightly more likely to be found at the ADE5,7 promoter than at the POL1 ORF. A similar observation was made by Kim et al. (18), who showed that the ratio of the HIS3/POL1 ChIP signal was only slightly above 1. We conclude that both SAGA and SWI/SNF have low-level association with the ADE5,7 promoter that is not increased under derepressing conditions.
FIG. 6.
SAGA and SWI/SNF occupy the ADE5,7 promoter independently of adenine. (A) ChIP assays were performed using antibodies to the myc tag on proteins Snf2, Spt7, and Bas1 along with an NTC strain, FY1353. A 500-fold dilution of the input control and the undiluted IP DNA were amplified by PCR by using primers specific for ADE5,7 and POL1 in the presence of [α-32P]dATP. Results from a representative ChIP are shown. The PCR products were resolved on 6% polyacrylamide gels, visualized, and quantified by phosphorimaging analysis. Enrichment (n-fold) was calculated as the ratios of the ADE5,7 signals in the IP-to-input samples and normalized for the corresponding ratios calculated for POL1 in the IP-to-input samples. (B) The resulting values from three ChIP experiments were plotted as the averages, with standard deviations, of the results from PCR amplifications of chromatin-immunoprecipitated DNA from three independent experiments. Strains grown with adenine are represented by open bars, and strains grown in medium lacking adenine are represented by filled bars. (C) Calculation of the percent IP was carried out by dividing the IP signal for ADE5,7 by the corresponding input signal. For all strains, values were significant, P ≤ 0.05, compared to the NTC. Bars are as defined for panel B.
SAGA and SWI/SNF association with the ADE5,7 promoter is independent of Bas1 and Pho2.
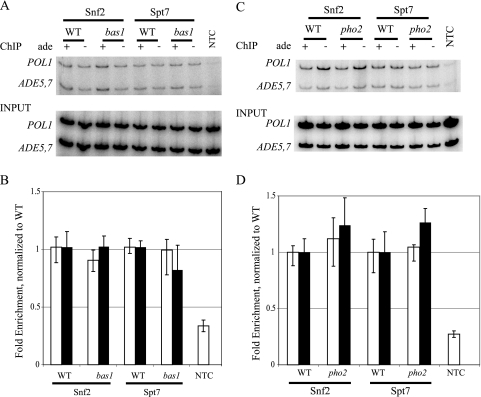
The association of SAGA and SWI/SNF at ADE5,7 might depend upon the transcriptional activator Bas1 or Pho2. We performed ChIP assays using an antibody to the myc tag on Spt7 and Snf2 in strains that carried null alleles of bas1 or pho2 (Fig. 7). Occupancy of SAGA and SWI/SNF was unchanged by the absence of either transcription factor.
FIG. 7.
SAGA and SWI/SNF associate with the ADE5,7 promoter independently of the Bas1 and Pho2 activators. (A) ChIPs of Spt7-myc and Snf2-myc were performed in wild-type and bas1Δ mutant strains (RR441 and RR443) along with an NTC strain, FY1353. (B) Graphical representation of the ChIP results. The ChIP data were plotted, showing the averages and standard deviations of results from PCR amplifications of chromatin immunoprecipitated from three independent experiments and normalized to wild-type levels. (C) ChIPs of Spt7-myc and Snf2-myc were performed in wild-type and pho2Δ mutant strains (RR442 and RR444). (D) Graphical representation of the averages and standard deviations of data from PCR amplifications of chromatin immunoprecipitated from three independent experiments and normalized to wild-type levels. WT, wild type. For panels B and D, open and filled bars represent strains grown in medium with and without adenine, respectively.
SAGA and SWI/SNF affect factor promoter occupancy of Bas1 and Pho2.
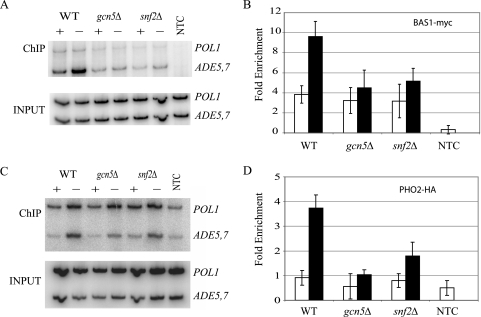
We explored the possibility that SAGA and SWI/SNF affect the binding of Bas1 or Pho2 to UASADE, altering their occupancy. Previous work from our laboratory, using ChIP assays, showed that Bas1 binds several ADE promoters under repressing conditions and its occupancy increases by approximately 2.2-fold under derepressing conditions (51). We used ChIP analysis to look for the presence of the Bas1 protein at the promoter of the ADE5,7 gene when strains harbored the gcn5Δ and snf2Δ deletion alleles. Interestingly, we found that the absence of Gcn5 or Snf2 affects the binding of Bas1-myc to ADE5,7 (Fig. 8A). Under the repressing conditions of SC medium containing adenine, Bas1 is present at the promoter in the mutants at the same level as that found in the wild type. However, under derepressing conditions of SC medium lacking adenine, binding of Bas1 to the promoter is clearly reduced in the mutants (Fig. 8A). These results indicate that the absence of SAGA and SWI/SNF activity decreases Bas1 occupancy at ADE5,7.
FIG. 8.
Promoter occupancy of Bas1 and Pho2 is decreased in SAGA and SWI/SNF mutants. (A) ChIPs of Bas1-myc were performed with wild-type (RR433), gcn5Δ (RR434), and snf2Δ (RR435) strains along with an NTC strain (FY1353). (B) Enrichment (n-fold) of Bas1 binding. ChIP assays were performed on strains grown with adenine (open bars) and without adenine (filled bars). PCR amplification from the ChIP experiments was quantified as the ratio of signal in the IP relative to input DNA, normalized to the POL1 signals. The average values from three independent ChIP experiments are plotted, with the standard deviations; mutants were significantly different from the wild type under derepressing conditions (P ≤ 0.05). (C) ChIPs of Pho2-HA were performed with wild-type (RR430), gcn5Δ (RR431), and snf2Δ (RR432) strains along with the NTC strain (FY1353). (D) Enrichment (n-fold) of Pho2 binding. ChIP analyses were prepared from strains grown with adenine (open bars) and without adenine (filled bars). PCR amplification from the ChIP experiments was quantified as the ratio of signal in the IP relative to input DNA, normalized to the POL1 signals. The average values from three independent ChIP experiments are plotted, with the standard deviations; mutants were significantly different from the wild type under derepressing conditions (P ≤ 0.05).
Similar results were observed with the binding of Pho2 to the ADE5,7 promoter. We used ChIP analysis to look for the presence of the Pho2 protein at the promoter of the ADE5,7 gene when strains harbored the gcn5Δ and snf2Δ deletion alleles. We found that the absence of Gcn5 or Snf2 decreases the binding of Pho2-HA to ADE5,7 under derepressing conditions but not under repressing conditions (Fig. 8B). The reduction in Pho2 binding could be direct or indirect through the decreased binding of Bas1, as we have shown elsewhere that Pho2 is unable to efficiently bind to the promoter in the absence of Bas1 (51). However, clearly the occupancy of this protein is affected by the loss of Gcn5 and Snf2.
Together, these results demonstrate a role for the SAGA and SWI/SNF complexes in facilitating wild-type levels of occupancy of the Bas1 and Pho2 transcription factors at the promoters under derepressing conditions but with little to no effect under repressing conditions. In the absence of the remodeling complexes, formation of the transcriptionally active Bas1-Pho2 heterodimer is reduced, with effects on the expression of the target ADE genes. However, SAGA and SWI/SNF are not apparently recruited by these activators.
ADE gene regulation requires SAGA and SWI/SNF catalytic activity.
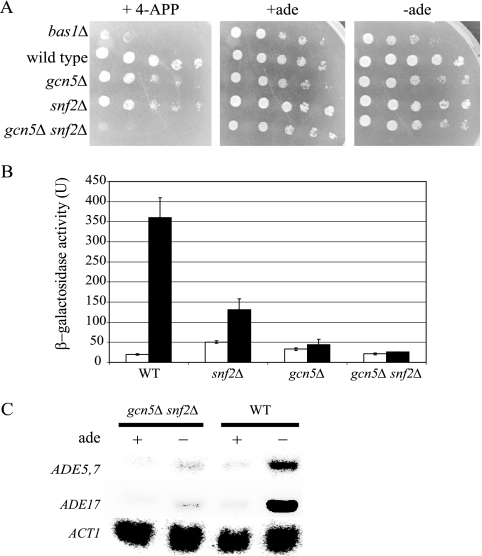
Our results indicate that both SAGA and SWI/SNF are necessary for ADE gene transcription; however, disruption of either complex alone lowered but did not completely eliminate ADE gene expression. Therefore, we hypothesized that the loss of both Gcn5 and Snf2 would exacerbate the reduction of ADE gene expression below that observed in either of the single mutants. We obtained a strain carrying mutations in both gcn5 and snf2 from Fred Winston (41); the mutant showed that the combined loss of the two genes resulted in very poor growth even on rich medium. We examined the sensitivity of the gcn5Δ snf2Δ strain to the adenine analog 4-APP to determine if sensitivity was increased relative to the single mutant strains. As shown in Fig. 9, each of the single gcn5Δ and snf2Δ mutants in this new strain background showed significant sensitivity to 4-APP compared to the wild type. Interestingly, the double mutant strain was extremely sensitive to 4-APP, much more so than either of the single mutants and about as sensitive as the bas1Δ strain (Fig. 9A). This finding suggests additive roles of the SAGA and SWI/SNF remodeling activities at the ADE genes.
FIG. 9.
gcn5 and snf2 are nonredundant for ADE transcription. (A) Wild-type, single mutant (gcn5Δ or snf2Δ), and double mutant (gcn5Δ snf2Δ) strains were grown to equivalent cell densities; serially diluted (1:10); spotted onto SC medium, SC medium containing 4-APP, and SC medium lacking adenine; and allowed to grow for approximately 48 h at 30°C. (B) The parental wild-type and the single and double mutant strains were transformed with a UASADE5,7-lacZ reporter (pR133). Strains were grown overnight in SC medium, inoculated into SC medium containing (open bars) or lacking (filled bars) adenine, and assayed for β-galactosidase activity. β-Galactosidase activities were measured in duplicate assays on at least three individual isolates. (C) Expression of ADE5,7 and ADE17 in parental wild-type, gcn5Δ and snf2Δ single mutant, and gcn5Δ snf2Δ double mutant strains was measured by Northern analysis. RNA was prepared from the four strains that had been grown in SC medium and transferred to SC medium lacking adenine. Expression was normalized to ACT1 transcript levels, and the experiment was repeated to confirm results.
We confirmed that this sensitivity to 4-APP was correlated with a loss of expression of the UASADE5,7-lacZ reporter (65). We found that both single mutant strains in this background were unable to fully derepress transcription (Fig. 9B). The snf2Δ mutation reduced transcription to approximately 35% of that in the isogenic wild type, whereas the gcn5Δ mutation had a stronger effect, reducing transcription to 10% of the wild-type level. The effect on gene expression in this strain background was more severe than what we had detected in the Research Genetics diploid knockout strain (Fig. 2). Interestingly, transcription in the gcn5Δ snf2Δ double mutant was barely detectable and virtually the same as that found under the repressing condition. Thus, the loss of catalytic activity from both complexes eliminated the ability to derepress the ADE5,7 gene. We obtained the same result using alternate ADE5,7-lacZ and ADE4-lacZ reporters (pR109 and pR115; data not shown): loss of either catalytic subunit alone decreased expression under derepressing conditions, but the loss of both activities resulted in only basal expression and the elimination of derepression.
To confirm the result from the lacZ reporter assays, we assessed expression from the chromosomal ADE5,7 and ADE17 loci in wild-type and mutant strains using Northern analysis. For each gene assayed, transcripts were barely detectable in double mutant strains (Fig. 9C). This result was consistent with the lacZ reporter expression. These results indicate that both the SAGA and SWI/SNF complexes are required for wild-type levels of ADE gene expression.
DISCUSSION
Genome-wide studies using DNA microarrays have investigated the loss of activity of the SAGA and SWI/SNF complexes. In one study, Holstege and colleagues (15) investigated the loss of both complexes from cells grown in rich (yeast extract-peptone-dextrose) medium, a condition in which the ADE regulon is repressed. Not surprisingly, their data showed that there was little to no difference in expression of the ADE regulon genes in the gcn5 and snf2 mutants relative to the wild type under these repressing conditions. Sudarsanam and colleagues (55) compared gene expression from cells grown in minimal medium with that from cells grown in rich medium to identify additional genes dependent upon SWI/SNF. About 1% of the genome showed an effect of threefold or greater. A great many genes, including several ADE genes, had small differences of less than threefold in gene expression in the snf2 and swi1 mutants. While these expression effects are small, they might nonetheless be significant under certain growth conditions.
In this report we demonstrate that the SAGA and SWI/SNF complexes are required for full expression of the ADE4, ADE5,7, and ADE17 genes, members of the ADE regulon. These remodeling complexes were identified by a genome-wide screen for 4-APP sensitivity, a phenotype exhibited by bas1Δ and pho2Δ mutants (Fig. 1) (Rolfes et al., unpublished), suggesting a role for the remodeling complexes in the expression of ADE genes. In some instances, the recruitment of the SAGA complex and its Gcn5 bromodomain have been shown to stabilize the SWI/SNF complex at promoter nucleosomes (13); thus, it was striking that the screen identified both the SAGA and SWI/SNF complexes. Indeed, the SAGA and SWI/SNF mutant strains exhibited decreased gene expression from several ADE genes in lacZ reporter and Northern assays (Fig. 2 and 3). Our data are consistent with the data from the Winston laboratory (55) in terms of the magnitude of the effect detected on transcription. Notably, however, we found that this difference in expression due to the loss of SAGA and SWI/SNF imposed a biological effect, namely, a lower tolerance for nucleotide pool imbalances.
Our data also indicate that the SAGA and SWI/SNF complexes are performing nonredundant roles in ADE gene expression. The double deletion strain was unable to derepress expression to any significant level above basal expression at ADE5,7-lacZ (Fig. 9B) or at native ADE5,7 and ADE17 loci (Fig. 9C). Thus, activity from either SAGA or SWI/SNF is necessary for a minimum derepression but full expression required each complex. We conclude that these complexes are performing complementary roles in modifying and remodeling chromatin necessary for ADE gene expression.
However, it is interesting that we observed virtually no 4-APP sensitivity in the ubp8, sgf11, and sgf73 mutant strains. This finding suggests that the activities performed by these components, such as deubiquitination, may not be critical for ADE gene expression. Alternatively, perhaps these subunits are not required for global surveillance (discussed below).
Association of the Bas1 and Pho2 activators with UASADE5,7 requires chromatin remodelers.
Many well-studied activators, such as Gcn4, have been shown to recruit the SAGA and SWI/SNF remodeling complexes during transcriptional activation (9). Less studied is the role of these complexes in the binding of transcription factors to their binding sites (32). We found that the SAGA complex (via Spt7-myc) and the SWI/SNF complex (via Snf2-myc) associate with the ADE5,7 promoter at a low but significant level that was surprisingly not affected by the SAICAR/AICAR regulatory signal or the presence of the activators Bas1 and Pho2. However, we found that an increase in Bas1 and Pho2 binding that occurs under derepressing conditions was dependent on SAGA and SWI/SNF. This result was strengthened by the observation that a constitutively active Bas1-Pho2 fusion protein also required the remodeling complexes.
Our data show that the SAGA and SWI/SNF remodeling complexes are not recruited by the Bas1 or Pho2 activator, yet these remodelers associate with the ADE5,7 gene and their activity is important for the increased binding of the activators and for transcription. This finding agrees with models for the action of the remodeling complexes described by other investigators. Cosma et al. (2) demonstrated that binding of SBF (Swi4/Swi6) required the activity of both SAGA and SWI/SNF whereas the binding of Swi5 did not. The association of Pho4 with nucleosomal UAS2 at PHO5 also required both Snf2 and Gcn5, although Pho4 binding at UAS1 did not require these remodelers (6, 52). Interestingly, both Swi5 and Pho4 cooperate with Pho2, but neither of these previous studies examined effects on Pho2 binding. We found a dependence on both SAGA and SWI/SNF for the binding of Pho2. While Bas1, Pho4, and Swi5 can all bind to their UAS elements in the absence of the remodeling complexes (presumably once Pho4 and Swi5 become nucleus localized), only Bas1 requires the remodelers to respond to the regulatory signal.
Both SAGA and SWI/SNF catalytic functions are important for ADE gene derepression.
Our data point to a second role for the chromatin remodeling complexes in transcription. We found that transcription driven by the Bas1-Pho2 fusion protein depended on both Gcn5 and Snf2. Normally, Bas1 and Pho2 do not interact until the regulatory signal is generated; combining the genes to produce the fusion protein makes reception of the signal moot. The Bas1-Pho2 fusion protein promotes transcription constitutively at the ADE1 (36) and ADE5,7 and ADE4 (Fig. 5) genes, indicating that it is constitutively bound to DNA, and yet it still requires the remodeling activities from both SAGA and SWI/SNF. Since a change in promoter occupancy under these conditions is unlikely, our data suggest that SAGA and SWI/SNF are involved at events downstream of factor binding, possibly remodeling chromatin for the binding of the general transcription factors or for transcriptional elongation. Further study will be necessary to define any additional roles.
Global surveillance.
How might the SAGA and SWI/SNF complexes associate with the ADE5,7 promoter since they are not recruited by Bas1 and Pho2? One possibility is that the complexes are recruited by another factor. Dhasarathy and Kladde (6) showed that myc-tagged Snf2 and Gcn5 are recruited to RPS22B by Abf1, even though the remodelers are not required for expression of this locus. It is possible that Abf1, which binds to the ADE5,7 promoter (44), recruits SAGA and SWI/SNF. However, since Abf1 is found only in the ADE5,7 promoter and not in the promoters for other ADE genes such as ADE4 and ADE17 (25, 44, 63), this putative mechanism would not explain the transcriptional requirement for SAGA and SWI/SNF at these other ADE genes. Perhaps other factors not studied in this report facilitate or enhance SWI/SNF and SAGA association, as was observed for Mediator at the ARG1 promoter (64).
Another explanation for the presence of SAGA and SWI/SNF at promoters is global surveillance. Through the global acetylation mechanism, HATs enzymatically modify histone proteins within large chromatin domains, including both coding sequences and intergenic regions, without any apparent DNA sequence specificity or specific recruitment. Kuo et al. (20) demonstrated a global role for the activity of the Gcn5 HAT. They showed that the level of H3 acetylation at many genes, including ADE5,7, was decreased in a gcn5 mutant, relative to the wild type, when the Gcn5-containing complexes were not specifically targeted to these promoters. Other researchers have also described a similar phenomenon for the Gcn5 HAT (16, 20, 60).
Our data are consistent with this global role, not only for Gcn5-containing complexes but also for the first time for the SWI/SNF complex. Thus, it is possible that a global surveillance role, while not fully understood, could explain the dependence on these chromatin remodeling complexes for ADE gene transcription.
SAGA and SWI/SNF function as coactivators in ADE gene derepression.
Our current model is that the presence of SAGA and SWI/SNF at the promoter facilitates efficient binding of the activators, Bas1 and Pho2, and thereby they function as coactivators in the derepression of ADE genes. SAGA and SWI/SNF are associated with ADE gene promoters as a part of their global surveillance role; we propose that they are present to efficiently and quickly turn on the ADE genes upon reception of the regulatory signal. There are other remodeling complexes previously shown to be poised under repressive conditions. NuA4, necessary for PHO5 activation and chromatin remodeling, is critical for chromatin remodeling even prior to activation, demonstrating that NuA4 readies or poises the PHO5 for activation (29). NuA4 is also present under repressing conditions, and, surprisingly, no increase in its association with promoters was detected upon induction. Interestingly, NuA4 was not actually required once the PHO5 gene was activated; Nourani et al. proposed that targeted hyperacetylation of nucleosomal histones under repressive conditions prepared the promoter and potentiated a rapid remodeling step, followed by transcriptional induction in response to signals (29). Other researchers have proposed that chromatin remodelers are continuously required because there is rapid, dynamic equilibrium between active and repressive chromatin structures (6). This is a possibility for SAGA and SWI/SNF involvement in ADE gene activation as well, where ADE genes need to be turned on quickly and efficiently. This “poising” and the continuous presence of the complexes at low levels of binding, as suggested by other researchers (29), are more efficient for stimulating gene expression quickly and thus rapidly producing biosynthetic products.
SAGA and SWI/SNF may therefore associate and be poised at low levels to facilitate the first round of transcription and the increase in factor occupancy. We also cannot rule out the possibility that SAGA and SWI/SNF association will not increase at later time points after induction, not detected at the initial rounds of transcription as well. Interestingly, Dhasarathy and Kladde observed that as late as 12 h after induction the occupancy of the activator Pho4 and the coactivators SAGA and SWI/SNF at the PHO5 promoters reached the maximum (6). How this increased binding at later times would correlate with gene expression is not inherently clear. Remodelers might be required not only for initial opening of chromatin but also for the long-term stabilization of the preinitiation complex, allowing for repeated RNA polymerase II loading.
Acknowledgments
This work was supported by MCB-0344371 from the National Science Foundation and institutional support from the Graduate School of Arts and Sciences of Georgetown University.
We are very grateful to Fred Winston for providing yeast strains FY1352, FY1353, FY1354, and FY1360. We also thank the Hinnebusch lab for HQY367 and HQY453 as well as Chhabi Govind for his assistance with ChIP assays. We thank David Clark for myc antibody and protein G-Dynabeads. We thank Odd Stokke Gabrielsen for the Bas1-Pho2 fusion-encoding plasmid.
Footnotes
Published ahead of print on 15 June 2007.
REFERENCES
- 1.Arndt, K. T., C. Styles, and G. R. Fink. 1987. Multiple global regulators control HIS4 transcription in yeast. Science 237:874-880. [DOI] [PubMed] [Google Scholar]
- 2.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 3.Daignan-Fornier, B., and G. R. Fink. 1992. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc. Natl. Acad. Sci. USA 89:6746-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denis, V., H. Boucherie, C. Monribot, and B. Daignan-Fornier. 1998. Role of the Myb-like protein Bas1p in Saccharomyces cerevisiae: a proteome analysis. Mol. Microbiol. 30:557-566. [DOI] [PubMed] [Google Scholar]
- 5.Denis, V., and B. Daignan-Fornier. 1998. Synthesis of glutamine, glycine and 10-formyl tetrahydrofolate is coregulated with purine biosynthesis in Saccharomyces cerevisiae. Mol. Gen. Genet. 259:246-255. [DOI] [PubMed] [Google Scholar]
- 6.Dhasarathy, A., and M. P. Kladde. 2005. Promoter occupancy is a major determinant of chromatin remodeling enzyme requirements. Mol. Cell. Biol. 25:2698-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gedvilaite, A., and K. Sasnauskas. 1994. Control of the expression of the ADE2 gene of the yeast Saccharomyces cerevisiae. Curr. Genet. 25:475-479. [DOI] [PubMed] [Google Scholar]
- 8.Giani, S., M. Manoni, and D. Breviario. 1991. Cloning and transcriptional analysis of the ADE6 gene of Saccharomyces cerevisiae. Gene 107:149-154. [DOI] [PubMed] [Google Scholar]
- 9.Govind, C. K., F. Zhang, H. Qiu, K. Hofmeyer, and A. G. Hinnebusch. 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25:31-42. [DOI] [PubMed] [Google Scholar]
- 10.Gregory, P. D., A. Schmid, M. Zavari, M. Munsterkotter, and W. Horz. 1999. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 18:6407-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guarente, L., and M. Ptashne. 1981. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78:2199-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannum, C., O. I. Kulaeva, H. Sun, J. L. Urbanowski, A. Wendus, D. J. Stillman, and R. J. Rolfes. 2002. Functional mapping of Bas2—identification of activation and Bas1-interaction domains. J. Biol. Chem. 277:34003-34009. [DOI] [PubMed] [Google Scholar]
- 13.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104:817-827. [DOI] [PubMed] [Google Scholar]
- 14.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 15.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 16.Imoberdorf, R. M., I. Topalidou, and M. Strubin. 2006. A role for Gcn5-mediated global histone acetylation in transcriptional regulation. Mol. Cell. Biol. 26:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingvarsdottir, K., N. J. Krogan, N. C. Emre, A. Wyce, N. J. Thompson, A. Emili, T. R. Hughes, J. F. Greenblatt, and S. L. Berger. 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25:1162-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, Y., N. McLaughlin, K. Lindstrom, T. Tsukiyama, and D. J. Clark. 2006. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol. Cell. Biol. 26:8607-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269-272. [DOI] [PubMed] [Google Scholar]
- 20.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 21.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 22.Lucchini, G., A. G. Hinnebusch, C. Chen, and G. R. Fink. 1984. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mantsala, P., and H. Zalkin. 1984. Glutamine nucleotide sequence of Saccharomyces cerevisiae ADE4 encoding phosphoribosylpyrophosphate amidotransferase. J. Biol. Chem. 259:8478-8484. [PubMed] [Google Scholar]
- 24.Mieczkowski, P. A., M. Dominska, M. J. Buck, J. L. Gerton, J. D. Lieb, and T. D. Petes. 2006. Global analysis of the relationship between the binding of the Bas1p transcription factor and meiosis-specific double-strand DNA breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:1014-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyake, T., J. Reese, C. M. Loch, D. T. Auble, and R. Li. 2004. Genome-wide analysis of ARS (autonomously replicating sequence) binding factor 1 (Abf1p)-mediated transcriptional regulation in Saccharomyces cerevisiae. J. Biol. Chem. 279:34865-34872. [DOI] [PubMed] [Google Scholar]
- 26.Mohrmann, L., and C. P. Verrijzer. 2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 1681:59-73. [DOI] [PubMed] [Google Scholar]
- 27.Neef, D. W., and M. P. Kladde. 2003. Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol. Cell. Biol. 23:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimura, K., K. Yasumura, K. Igarashi, S. Harashima, and Y. Kakinuma. 1999. Transcription of some PHO genes in Saccharomyces cerevisiae is regulated by Spt7p. Yeast 15:1711-1717. [DOI] [PubMed] [Google Scholar]
- 29.Nourani, A., R. T. Utley, S. Allard, and J. Cote. 2004. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 23:2597-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Martin, J., and A. D. Johnson. 1998. Mutations in chromatin components suppress a defect of Gcn5 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1049-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson, C. L., A. Dingwall, and M. P. Scott. 1994. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. USA 91:2905-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 33.Pickering, W. R., and R. A. Woods. 1972. Altered purine permease activity in a purine-analogue-resistant mutant of Saccharomyces cerevisiae. Biochem. J. 127:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickering, W. R., and R. A. Woods. 1973. Genetics of resistance to 4-aminopyrazolo-(3,4-d)-pyrimidine in Saccharomyces cerevisiae. Mol. Gen. Genet. 122:231-242. [DOI] [PubMed] [Google Scholar]
- 35.Pickering, W. R., and R. A. Woods. 1972. The uptake and incorporation of purines by wild-type Saccharomyces cerevisiae and a mutant resistant to 4-aminopyrazolo (3,4-d) pyrimidine. Biochim. Biophys. Acta 264:45-58. [DOI] [PubMed] [Google Scholar]
- 36.Pinson, B., T. L. Kongsrud, E. Ording, L. Johansen, B. Daignan-Fornier, and O. S. Gabrielsen. 2000. Signaling through regulated transcription factor interaction: mapping of a regulatory interaction domain in the Myb-related Bas1p. Nucleic Acids Res. 28:4665-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell, D. W., C. M. Weaver, J. L. Jennings, K. J. McAfee, Y. He, P. A. Weil, and A. J. Link. 2004. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell. Biol. 24:7249-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu, H., C. Hu, F. Zhang, G. J. Hwang, M. J. Swanson, C. Boonchird, and A. G. Hinnebusch. 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 25:3461-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rebora, K., C. Desmoucelles, F. Borne, B. Pinson, and B. Daignan-Fornier. 2001. Yeast AMP pathway genes respond to adenine through regulated synthesis of a metabolic intermediate. Mol. Cell. Biol. 21:7901-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rebora, K., B. Laloo, and B. Daignan-Fornier. 2005. Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: a central role for a small molecule. Genetics 170:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts, S. M., and F. Winston. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147:451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rolfes, R. J. 2006. Regulation of purine nucleotide biosynthesis: in yeast and beyond. Biochem. Soc. Trans. 34:786-790. [DOI] [PubMed] [Google Scholar]
- 43.Rolfes, R. J., and A. G. Hinnebusch. 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13:5099-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rolfes, R. J., F. Zhang, and A. G. Hinnebusch. 1997. The transcriptional activators BAS1, BAS2, and ABF1 bind positive regulatory sites as the critical elements for adenine-regulation of ADE5,7. J. Biol. Chem. 272:13343-13354. [DOI] [PubMed] [Google Scholar]
- 45.Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194:281-301. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 47.Sherman, F., G. R. Fink, and C. W. Lawrence. 1974. Methods of yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 48.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, C. L., R. Horowitz-Scherer, J. F. Flanagan, C. L. Woodcock, and C. L. Peterson. 2003. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 10:141-145. [DOI] [PubMed] [Google Scholar]
- 50.Smith, C. L., and C. L. Peterson. 2005. A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol. Cell. Biol. 25:5880-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Som, I., R. N. Mitsch, J. L. Urbanowski, and R. J. Rolfes. 2005. DNA-bound Bas1 recruits Pho2 to activate ADE genes in Saccharomyces cerevisiae. Eukaryot. Cell 4:1725-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steger, D. J., E. S. Haswell, A. L. Miller, S. R. Wente, and E. K. O'Shea. 2003. Regulation of chromatin remodeling by inositol polyphosphates. Science 299:114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stotz, A., P. P. Muller, and P. Linder. 1993. Regulation of the ADE2 gene from Saccharomyces cerevisiae. Curr. Genet. 24:472-480. [DOI] [PubMed] [Google Scholar]
- 55.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swanson, M. J., H. Qiu, L. Sumibcay, A. Krueger, S. J. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23:2800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Syntichaki, P., I. Topalidou, and G. Thireos. 2000. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 404:414-417. [DOI] [PubMed] [Google Scholar]
- 58.Tice-Baldwin, K., G. R. Fink, and K. T. Arndt. 1989. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science 246:931-935. [DOI] [PubMed] [Google Scholar]
- 59.Valerius, O., C. Brendel, C. Wagner, S. Krappmann, F. Thoma, and G. H. Braus. 2003. Nucleosome position-dependent and -independent activation of HIS7 epression in Saccharomyces cerevisiae by different transcriptional activators. Eukaryot. Cell. 2:876-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 61.Voth, W. P., Y. W. Jiang, and D. J. Stillman. 2003. New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast 20:985-993. [DOI] [PubMed] [Google Scholar]
- 62.Wu, P. Y., C. Ruhlmann, F. Winston, and P. Schultz. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15:199-208. [DOI] [PubMed] [Google Scholar]
- 63.Yarragudi, A., L. W. Parfrey, and R. H. Morse. 2007. Genome-wide analysis of transcriptional dependence and probable target sites for Abf1 and Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 35:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoon, S., H. Qiu, M. J. Swanson, and A. G. Hinnebusch. 2003. Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol. Cell. Biol. 23:8829-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, F., M. Kirouac, N. Zhu, A. G. Hinnebusch, and R. J. Rolfes. 1997. Evidence that complex formation by Bas1p and Bas2p (Pho2p) unmasks the activation function of Bas1p in an adenine-repressible step of ADE gene transcription. Mol. Cell. Biol. 17:3272-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]