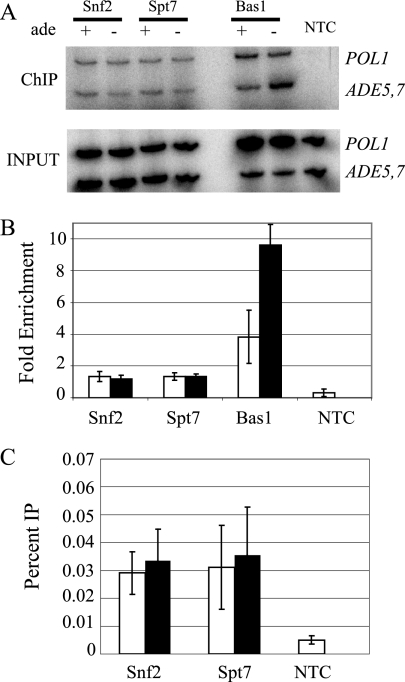
FIG. 6.
SAGA and SWI/SNF occupy the ADE5,7 promoter independently of adenine. (A) ChIP assays were performed using antibodies to the myc tag on proteins Snf2, Spt7, and Bas1 along with an NTC strain, FY1353. A 500-fold dilution of the input control and the undiluted IP DNA were amplified by PCR by using primers specific for ADE5,7 and POL1 in the presence of [α-32P]dATP. Results from a representative ChIP are shown. The PCR products were resolved on 6% polyacrylamide gels, visualized, and quantified by phosphorimaging analysis. Enrichment (n-fold) was calculated as the ratios of the ADE5,7 signals in the IP-to-input samples and normalized for the corresponding ratios calculated for POL1 in the IP-to-input samples. (B) The resulting values from three ChIP experiments were plotted as the averages, with standard deviations, of the results from PCR amplifications of chromatin-immunoprecipitated DNA from three independent experiments. Strains grown with adenine are represented by open bars, and strains grown in medium lacking adenine are represented by filled bars. (C) Calculation of the percent IP was carried out by dividing the IP signal for ADE5,7 by the corresponding input signal. For all strains, values were significant, P ≤ 0.05, compared to the NTC. Bars are as defined for panel B.