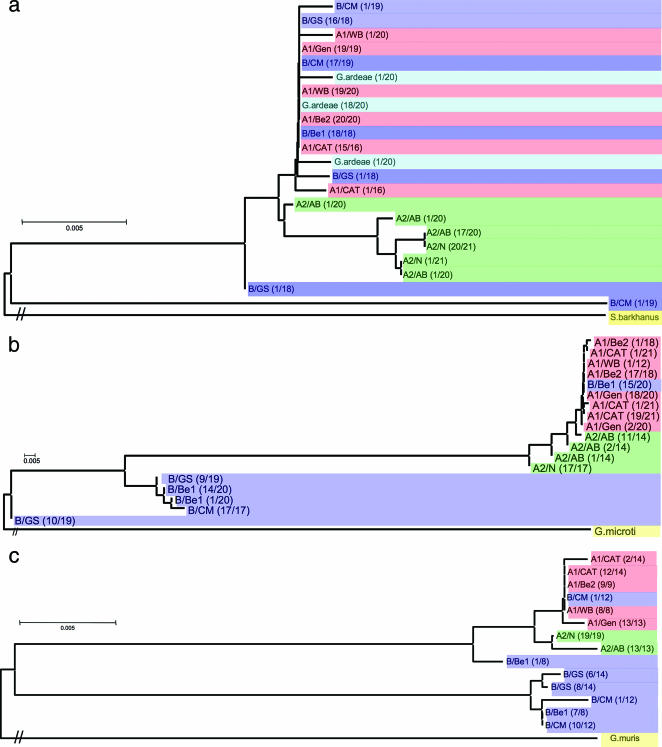
FIG. 1.
Neighbor-joining phylogenetic analyses of nine Giardia lamblia isolates at (a) actin, (b) TPI, and (c) beta giardin loci, carried out using the Kimura two-parameter model. Each unique sequence was used only once in the construction of the trees; thus, most branches represent multiple identical sequences. The haplotypes were named as follows: group designation/isolate designation. The frequency of each haplotype is shown in parentheses. Spironucleus barkhanus (GenBank accession number DX916114), Giardia microti (GenBank accession number AY228649), and Giardia muris (GenBank accession number AY258618) sequences were set as outgroups in the analyses of actin, TPI, and beta giardin loci, respectively. Different colors illustrate distinct groups: pink for group A-1, green for group A-2, blue for group B, light blue for G. ardeae, and yellow for the outgroup. For topology purposes, hash marks were used on the outgroup branches because of the extremely long branch lengths. Two major groupings are first apparent: A-1/B/A-2 and B. The first group is further subdivided into A-1/B and A-2. The trees show the presence of divergent haplotypes (A-1/B and B-specific haplotypes) in group B sequences. The group B-specific haplotype is the rare one at the actin locus but the prevalent one at the TPI and beta giardin loci.