Abstract
The oral pathogen Porphyromonas gingivalis secretes proteases such as Arg-gingipain B (RgpB) that activate protease-activated receptors (PARs). Human beta-defensins (hBDs) and the macrophage inflammatory protein 3α/CC chemokine ligand 20 (CCL20) produced by epithelial cells are antimicrobial peptides that provide cytokine function and play an important role in innate immunity. The aim of the present study was to determine whether specific members of the PAR family mediate the expression of these innate immunity markers in gingival epithelial cells (GECs) when exposed to P. gingivalis cell-free culture supernatant or purified RgpB. hBD-2 mRNA in GECs was induced in response to supernatant and purified RgpB from P. gingivalis (P = 0.02 and P = 0.016, respectively). This effect was abrogated by the protease inhibitor tosyl-l-lysine chloromethyl ketone (TLCK) (P < 0.05). In response to P. gingivalis supernatant and to purified RgpB, the hBD-2 mRNA expression was significantly decreased in PAR-2 gene knockdown cells, whereas no change was detected in PAR-1 gene knockdown cells. CCL20 mRNA expression also increased in response to the supernatant of P. gingivalis, and this effect was blocked by the protease inhibitor, TLCK (P = 0.05 and P = 0.024, respectively), and was blocked in PAR-2 gene knockdown cells. Our data indicate that hBD-2 and CCL20 mRNA up-regulation by P. gingivalis supernatant and purified RgpB was mediated via PAR-2, but not via PAR-1, and that proteases play a role in the regulation of innate immune responses in GECs. GECs use PARs to recognize P. gingivalis and mediate cell responses involved in innate immunity.
The oral epithelium is exposed to large numbers of commensal and pathogenic microorganisms, requiring an efficient ecologic balance between microbiologic colonization and the local immune response. Epithelial cell recognition of bacteria and the distinction between commensal and pathogenic bacteria are important processes that occur in the gingival epithelium. It is widely accepted that cells utilize pattern recognition receptors to identify bacteria in their environment (19, 51), but in addition, some pathogens, such as Porphyromonas gingivalis, secrete proteases that are recognized by cells via the family of protease-activated receptors (41). Recently, it has been shown that protease-activated receptor 2 (PAR-2) is involved in inflammatory processes in several tissues (24, 25, 60).
PARs are a family of G protein-coupled, seven-transmembrane-domain receptors that mediate various cellular responses to proteases such as thrombin, trypsin, and mast cell tryptase (21, 26). PARs are activated by proteolytic cleavage of the N-terminal domain by extracellular proteases. This process reveals their N-terminal “tethered ligand,” which leads to intracellular signaling (5, 39). Four PAR family members (PAR-1, -2, -3, and -4) have been identified so far. While PAR-1, -3, and -4 are activated by thrombin and are involved in platelet aggregation, PAR-2 is activated by trypsin, mast cell tryptase, neutrophil protease 3, tissue factor/factor VIIa/factor Xa, and membrane-tethered serine protease 1 (26, 40, 58). Although PARs are widely expressed, PAR-1 and PAR-2 are the main PARs found in the epithelium of the gastrointestinal tract (43) and gingiva (40).
P. gingivalis is a gram-negative, obligately anaerobic bacterium that is a major etiologic factor in the development of chronic periodontitis (23, 37). The gingipains are proteases that are synthesized by P. gingivalis and have been recognized as crucial virulence factors. Gingipains are involved in the degradation of the adherens junctions between cells, which might allow P. gingivalis to invade into the epithelium and to deeper tissues (1, 20, 36, 52, 53). In culture, three types of gingipains are secreted into the medium of P. gingivalis: Arg-gingipains A and B (RgpA and RgpB) and Lys-gingipain (Kgp) (48, 49). P. gingivalis and purified gingipains have been used to investigate activation of PARs in oral epithelial cells, which resulted in increased expression of the antimicrobial peptide human beta-defensin-2 (hBD-2) (4) and inflammatory cytokines (24, 40), respectively. PARs have been implicated in the pathogenesis of periodontal disease in an animal model (24, 25) and in a transfected epithelial cell model (24).
hBDs are antimicrobial peptides present in the chemical barrier as a part of the innate immunity provided by epithelial cells (13, 45). hBDs are small (<100-amino-acid), polycationic beta-sheet molecules that possess a broad spectrum of activity against both gram-negative and gram-positive bacteria and some fungi and viruses (15, 38). hBD-2 is an inducible antimicrobial peptide (10, 16-18, 35, 44, 46) that shows strong bactericidal effects against gram-negative bacteria that are prevalent in periodontal disease (16, 22, 28).
Macrophage inflammatory protein 3α/CC chemokine ligand 20 (CCL20) is a chemokine with regions that are structurally related to hBD-2, and like hBD-2 it exhibits antimicrobial activity in vitro against Escherichia coli and Staphylococcus aureus (27). Both CCL20 and hBD-2 are comparable mediators linking the innate and adaptive immunities (55, 62). Both hBD-2 and CCL20 interact with chemokine receptor 6 and are chemoattractants for immature dendritic cells (55, 61).
Despite the fact that hBD-2 is considered an inducible peptide at sites of inflammation, it is expressed in healthy, clinically noninflamed gingival epithelium (7, 8). The oral commensal bacterium Fusobacterium nucleatum is known to induce gene expression of hBD-2 (35), which may be a partial explanation of this phenomenon. Our laboratory has shown that induction of hBD-2 by commensal bacteria is via mitogen-activated protein kinase and calcium signaling pathways, while induction by pathogenic bacteria is via mitogen-activated protein kinase and NF-κB signaling pathways (3, 33, 34). Because Toll-like receptors may not be involved in hBD-2 up-regulation in oral epithelial cells (35), other receptors, such as protease-activated receptors, have been suggested to be activated by oral pathogenic bacteria (4, 40).
We hypothesized that gingival epithelial cells utilize PAR-2 to mediate the gene expression of hBD-2 and CCL20 in response to proteases synthesized and secreted by P. gingivalis. Thus, we tested whether PAR-1, PAR-2, or both PAR-1 and PAR-2 are involved in the recognition of P. gingivalis by cultured gingival epithelial cells. It was further of interest to investigate the gene expression of hBD-2 and CCL20 in response to cell-free supernatant and purified RgpB from P. gingivalis in order to confirm the role of the secreted proteases independent from lipopolysaccharides (LPS) and the ability of the live P. gingivalis bacteria to invade epithelial cells.
We show evidence that the proteases secreted by P. gingivalis up-regulate hBD-2 and CCL20 mRNAs via PAR-2 and, further, that P. gingivalis has an effect on PAR-1 and PAR-2 gene expression which may enhance the overall effect of the gingipains on epithelial cells.
MATERIALS AND METHODS
Human gingival epithelial cells and cell transfection.
Gingival biopsies were obtained from healthy patients who underwent third-molar extraction in the Department of Oral Surgery, School of Dentistry, University of Washington, in accordance with a University of Washington Institutional Review Board-approved study. The tissue was prepared for cell culture as described previously by our group (4). Epithelial cells were cultured in keratinocyte growth medium with 0.15 mM Ca2+ using the supplements from the KGM-bullet kit (Cambrex, Walkersville, MD). The cells were cultured at 37°C in a humidified atmosphere (5% CO2).
For gene silencing, guaranteed small interfering RNA (siRNA) tagged with Alexa Fluor 488 (QIAGEN, Valencia, CA) was used to target the human PAR-1 or PAR-2 gene. The sequences and target locations are listed in Table 1. The fast-forward transfection protocol was performed according to the manufacturer's instructions. In brief, siRNA (25 nM) was mixed with HiPerFect reagent and ECR buffer (QIAGEN) to obtain the transfection complex, which was added to 5.5 × 104 cells at 1 h after seeding (24-well culture plate). Forty-eight hours later, the cells were stimulated as described below. Scrambled nonsilencing RNA served as a negative control and was transfected using the same concentration as for PAR siRNA. The lipid carrier HiPerFect was used as an additional control for all experiments. Transfection efficiency was monitored using a fluorescence microscope (Eclipse TS100; Nikon, Melville, NY) and confirmed by real-time PCR. The appropriate siRNA concentration was determined using different concentrations (10 nM, 25 nM, 50 nM, and 100 nM of siRNA) in preliminary experiments.
TABLE 1.
Sequences of siRNAs
| Target or siRNA | Sequence
|
|
|---|---|---|
| PAR-1 | PAR-2 | |
| Target | 5′-CAG TAT AGA ATA GGC ACT TTA-3′ | 5′-TAG GAT GTG GAA CTT GTT TAA-3′ |
| Sense siRNA | r(GUA UAG AAU AGG CAC UUU A)dTdT | r(GGA UGU GGA ACC UGU UUA A)dTdT |
| Antisense siRNA | r(UAA AGU GCC UAU UCU AUA C)dTdG | r(UUA AAC AGG UUC CAC AUC C)dTdA |
P. gingivalis culture conditions and treatment.
Wild-type P. gingivalis strain 33277 was cultured to the late logarithmic growth phase as described previously (4). Bacterial numbers were estimated by absorbance measurement using the TECAN GENios Multidetection Reader, V.4.51 (Phoenix, Hayward, CA). To obtain the supernatant, the bacterial cells were centrifuged at 1,200 × g for 5 min at 4°C. Subsequently, aliquots of the supernatant were used for preincubation (10 min) with 1 mmol/liter of the serine and cysteine protease inhibitor tosyl-l-lysine chloromethyl ketone (TLCK) (Sigma, St. Louis, MO), which inhibits the gingipains (12, 24). The protease inhibitor was diluted in endotoxin-free water (HyPure; HyClone, Logan, UT). The gingival epithelial cells were grown to 80% confluence and stimulated with either P. gingivalis supernatant or TLCK-preincubated supernatant, using an amount equivalent to a multiplicity of infection of 100:1, for 16 h. Blank medium served as a negative control for the stimulation experiments. Each stimulation experiment was performed in triplicate, and cells from two to five different donors were tested.
Purification of RgpB and treatment.
RgpB was purified as previously described by Rangarajan and coworkers (50). In brief, RgpB was purified from the growth medium of the deletion mutant of P. gingivalis W50 beg (PG1135 to PG1141), which produces only RgpA and RgpB. A high yield of RgpB can be obtained from this W50 beg strain, and this is identical to the RgpB described previously (50). The culture supernatant was centrifuged (10 000 × g, 60 min), and solid ammonium sulfate was added to the supernatant to 85% saturation to precipitate the Arg-X proteases. The protein was separated by centrifugation and suspended in 50 mM sodium acetate buffer (pH 5.3) containing 0.0055% Zwittergent (Calbiochem Novabiochem UK Ltd., Nottingham, United Kingdom). Insoluble material was separated by centrifugation, while the soluble, enzyme-containing fraction was subjected to gel filtration and finally affinity chromatography on Arg-agarose columns. This step was performed to separate RgpA and RgpB. Unbound RgpB was dialyzed against 50 mM sodium acetate buffer (pH 5.3) containing 0.0055% Zwittergent and purified by ion-exchange chromatography. Specific activity determination and sodium dodecyl sulfate-polyacrylamide gel electrophoresis were performed to evaluate pure enzyme fractions before dialysis against 50 mM sodium acetate buffer (pH 5.3) and 0.0055% (wt/vol) Zwittergent (50). The purified enzyme was stored at 4°C. RgpB was used at ∼2.5 units/ml, which is comparable to the activity of Arg-gingipains in P. gingivalis supernatants. For stimulation experiments, amounts of purified RgpB and P. gingivalis supernatant used in this study were comparable. Cells stimulated with TLCK-preincubated RgpB (10 min at room temperature) and heat-denatured RgpB (70°C, 10 min) served as controls. Each stimulation experiment was performed in triplicate overnight (16 h), and cells from two to five different donors were tested.
Conditions for RT-PCR and real-time PCR.
After stimulation, total RNA was extracted using the RNeasy minikit (QIAGEN). The reverse transcription reaction was performed using 500 ng of total RNA. The reaction mix contained 1× reverse transcriptase (RT) buffer, 250 nM oligo(dT) primer, 10 mM deoxynucleoside triphosphate mix, 50 U of RT, and 13 U of RNase inhibitor (Ambion, Austin, TX), and the reaction was carried out following standard protocols as previously described (4). Controls without RT enzyme were included with every experiment.
Quantitative analysis of the cDNA was performed using the MyiQiCycler (Bio-Rad, Hercules, CA) and Brilliant SYBR green PCR kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. PCRs were carried out in 96-well plates in a total volume of 25 μl, including 1 μl of cDNA and 250 nM primers (Table 2). At the end of every real-time PCR, melting curve analysis was performed to confirm that the amplified product was specific. Standard curve analysis was conducted, confirming a linear dependency (efficiency) between the cDNA concentration and the threshold cycle calculated by the iQ5 software (Bio-Rad). All reactions were carried out in duplicate, and average threshold cycle values were calculated. Sample values were normalized to the expression of the housekeeping ribosomal phosphoprotein gene, and relative expression was calculated using the mathematical model proposed by Pfaffl (3, 47). PCR controls were performed using water instead of cDNA. The data were statistically analyzed using the paired two-tailed t test (SPSS version 14). A P value of ≤0.05 was considered significant.
TABLE 2.
Oligonucleotide sequences used for real-time PCR
| Gene product | Oligonucleotide sequence
|
|
|---|---|---|
| Sense | Antisense | |
| PAR-1 | GTG ATT GGC AGT TTG GGT CT | GCC AGA CAA GTG AAG GAA GC |
| PAR-2 | CCT GGC CAT GTA CCT GAT CT | GAC ACT TCG GCA AAG GAG AG |
| hBD-2 | CCA GCC ATC AGC CAT GAG GGT | GGA GCC CTT TCT GAA TCC GCA |
| CCL20 | TTT ATT GTG GGC TTC ACA CG | GAT TTG CGC ACA CAG ACA AC |
| Ribosomal phosphoprotein | GCC TTG ACC TTT TCA GCA AG | GCA GCA TCT ACA ACC CTG AAG |
Protease assay.
The proteolytic activity of the P. gingivalis cell-free supernatant and purified RgpB was tested using the PDQ protease assay (AthenaES, Baltimore, MD) following the manufacturer's instructions. The supernatant and RgpB as well as the controls were prepared as described above. Test and control samples were prepared at a 2:1 dilution in buffer (100 mM phosphate-buffered saline [pH 7.2] with 5 mM cysteine and 5 mM EDTA) according to the manufacturer's suggestions. Triplicate reactions were carried out in a total volume of 0.5 ml. Papain (Sigma) was used as positive control, while buffer, TLCK, and blank bacterial medium served as negative controls. Subsequently, the samples were incubated at 37°C for 2, 4, 8, and 16 h. The enzyme reaction was terminated using a solution of 0.2 N NaOH (0.5 ml). Absorbance at 450 nm was analyzed using the TECAN reader.
RESULTS
PAR-1 and -2 mRNA expression is blocked by siRNA.
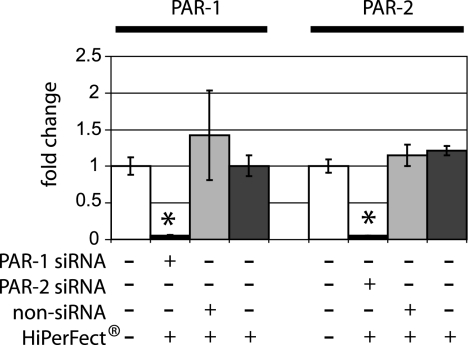
To block the gene expression of PAR-1 or PAR-2, human gingival epithelial cells were transfected with siRNA specific for each gene for 48 h. The intracellular uptake as well as the nuclear localization of the Alexa Fluor 488-labeled siRNA was documented by fluorescence at 24 and 48 h. Distinct intra- and perinuclear localizations of both PAR-1 and PAR-2 siRNA fluorescence signals were shown in over 95% of the transfected cells. Gene knockdown was confirmed by quantitative real-time PCR at 48 h after transfection. The mRNA expression of PAR-1 (P = 0.05) and PAR-2 (P = 0.03) was significantly decreased for all experimental groups compared to the controls (Fig. 1). Overall, at least a 90% PAR-1 and PAR-2 gene knockdown was demonstrated.
FIG. 1.
Effect of siRNA on PAR1 and PAR2 gene expression in gingival epithelial cells. Gingival epithelial cells were transfected with siRNA targeting PAR-1, PAR-2, or scrambled nonsilencing RNA. Efficiency of gene knockdown was evaluated by real-time PCR after 48 h. The gene expression of PAR-1 and PAR-2 was significantly lower than that in untransfected control cells (P = 0.05 and P = 0.03, respectively). Nonsilencing RNA and treatment with the lipid carrier HiPerFect did not influence either the PAR-1 or PAR-2 mRNA expression. The gene expression study was performed with triplicate samples derived from two to five different donors. *, significant difference (P ≤ 0.05). Error bars indicate standard deviations.
Proteolytic activity of P. gingivalis cell-free supernatant alters PAR-1 and PAR-2 gene expression.
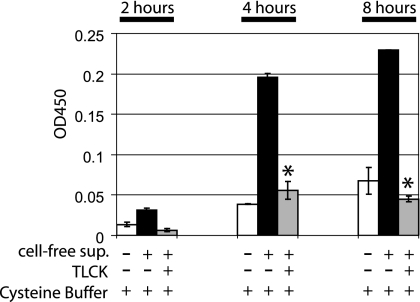
Proteolytic activity of the supernatant from P. gingivalis was demonstrated after 2, 4, and 8 h using a multisubstrate assay method. Preincubation of the supernatant with the protease inhibitor TLCK showed significantly reduced proteolytic activity compared to that in native supernatant after 4 and 8 h (P = 0.008 and P = 0.001, respectively) (Fig. 2).
FIG. 2.
Protease activity (PDQ protease activity assay) of native and TLCK-preincubated cell-free supernatants from P. gingivalis. The proteolytic activity of P. gingivalis supernatant was monitored for 2 h (data in duplicate), 4 h, and 8 h (data in triplicate). Native supernatant showed significantly higher proteolytic activity than TLCK-preincubated supernatant after 4 and 8 h (P = 0.008 and P = 0.001, respectively). Complete bacterial medium and TLCK served as negative controls and showed optical densities similar to that of the cysteine buffer (data not shown). Measurements were performed by absorbance at 450 nm, and the values were normalized to blank control samples. Error bars indicate standard deviations.
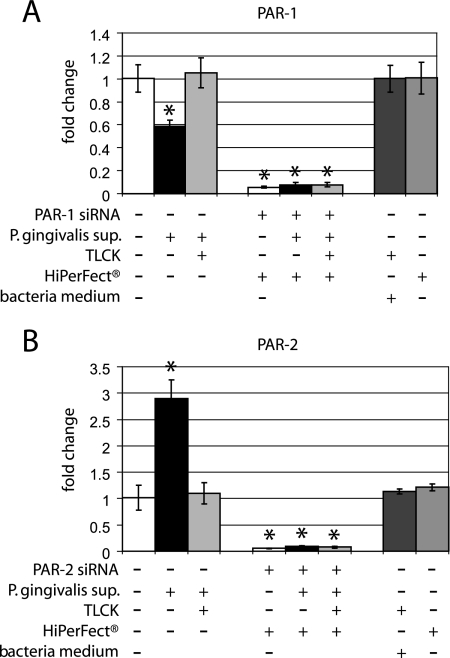
To evaluate the effect of the P. gingivalis cell-free supernatant on expression of PAR-1 and PAR-2 mRNAs, gingival epithelial cells were grown to 80% confluence and stimulated with the supernatant for 16 h. Analysis of gene expression using real-time PCR showed a down-regulation of PAR-1 mRNA in comparison to the untreated control cells (P = 0.05) (Fig. 3A). In contrast, the mRNA expression of PAR-2 was significantly up-regulated (P = 0.004) in response to P. gingivalis supernatant compared to that in unstimulated control cells (Fig. 3B). Preincubation of the P. gingivalis supernatant with the protease inhibitor TLCK abolished the effect and restored the PAR mRNA expression level to that of untreated cells. Controls using the transfection agent HiPerFect, or the blank bacterial medium, and TLCK showed no effect on the mRNA expression of PAR-1 or PAR-2. (Fig. 3A and B).
FIG. 3.
PAR-1 and PAR-2 gene expression in response to P. gingivalis supernatant in the presence and absence of siRNA. (A) PAR-1 gene expression was evaluated by real-time PCR in response to cell-free supernatant from P. gingivalis and TLCK-treated supernatant in untransfected cells and cells transfected with siRNA targeting PAR-1. Expression of PAR-1 mRNA was significantly down-regulated in response to P. gingivalis supernatant compared to that in unstimulated cells (P = 0.05). PAR-1 mRNA was significantly lower after stimulation with the supernatant as well as with TLCK-pretreated supernatant in all siRNA-transfected cells (P < 0.05) compared to untransfected stimulated and unstimulated gingival epithelial cells. Both controls, bacteria medium and the lipid carrier (HiPerFect), showed no effect on the gene expression of PAR-1. (B) The mRNA expression of PAR-2 was significantly up-regulated in response to cell-free supernatant from P. gingivalis compared to that in unstimulated cells (P = 0.004). PAR-2 mRNA was significantly lower after stimulation with the supernatant as well as with TLCK-pretreated supernatant in all siRNA-transfected cells (P < 0.05) compared to untransfected stimulated and unstimulated gingival epithelial cells. Both controls, bacterial medium and the lipid carrier (HiPerFect), showed no effect on the gene expression of PAR-2. The gene expression study was performed with triplicate samples derived from two to five different donors. *, significant difference (P ≤ 0.05). Error bars indicate standard deviations.
PAR-2 mediates hBD-2 mRNA expression in response to P. gingivalis cell-free supernatant.
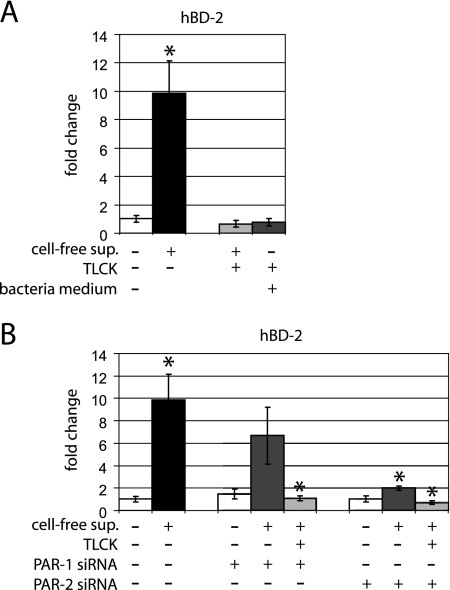
We previously showed that P. gingivalis up-regulated hBD-2 mRNA expression in gingival epithelial cells (4). To characterize this effect more completely, P. gingivalis cell-free supernatant was tested with and without the addition of TLCK and in cells in which the expression of PAR-1 or PAR-2 was blocked by siRNA. The gene expression of hBD-2 was significantly up-regulated in response to P. gingivalis cell-free supernatant (P = 0.02) compared to that in unstimulated gingival epithelial cells (Fig. 4A). Pretreatment of the supernatant with TLCK completely abrogated this effect (P = 0.024) (Fig. 4A), supporting the role of secreted proteases in hBD-2 regulation. The blank bacterial medium and TLCK controls did not alter mRNA expression of hBD-2 in comparison to unstimulated control cells.
FIG. 4.
hBD-2 mRNA expression in response to P. gingivalis supernatant and effects of siRNA for PAR-1 and PAR-2 in gingival epithelial cells. (A) The mRNA expression of hBD-2 was significantly up-regulated in response to the supernatant (P = 0.02). Stimulation of gingival epithelial cells with TLCK-pretreated supernatant and the control with complete bacterial media showed no significant change in hBD-2 mRNA expression compared to that in the unstimulated control. (B) The gene expression of hDB-2 was up-regulated in PAR-1 gene knockdown cells in response to the supernatant compared to that in untransfected unstimulated cells. No difference between nontransfected and transfected cells was demonstrated in response to P. gingivalis supernatant (P = 0.325). This effect was completely abolished by TLCK pretreatment (P = 0.025). In contrast, in PAR-2 gene knockdown cells, the mRNA expression of hBD-2 showed significantly lower levels in response to the supernatant compared to that in untransfected stimulated cells (P = 0.024). The gene expression study was performed with triplicate samples derived from two to five different donors. *, significant difference (P ≤ 0.05). Error bars indicate standard deviations.
The P. gingivalis cell-free supernatant also resulted in an up-regulation of hBD-2 mRNA in epithelial cells transfected with PAR-1 siRNA. This effect was blocked when the cell-free supernatant was preincubated with TLCK (P = 0.025) (Fig. 4B). In contrast, the hBD-2 mRNA was significantly lower (P = 0.024) in stimulated PAR-2 gene knockdown cells than in untransfected, supernatant-stimulated cells. The protease inhibitor TLCK significantly abolished the stimulatory effects of P. gingivalis supernatant on the mRNA expression profile of hBD-2 in both PAR-1 and -2 gene knockdown cells (P = 0.026 and P = 0.02, respectively) (Fig. 4B). Controls using the transfection agent HiPerFect did not affect hBD-2 gene expression (data not shown). The magnitude of hBD-2 gene expression varied in cells from different donors, but the overall pattern of effects of P. gingivalis supernatant and siRNA gene knockdown was consistent.
PAR-2 mediates CCL20 mRNA expression in response to P. gingivalis cell-free supernatant.
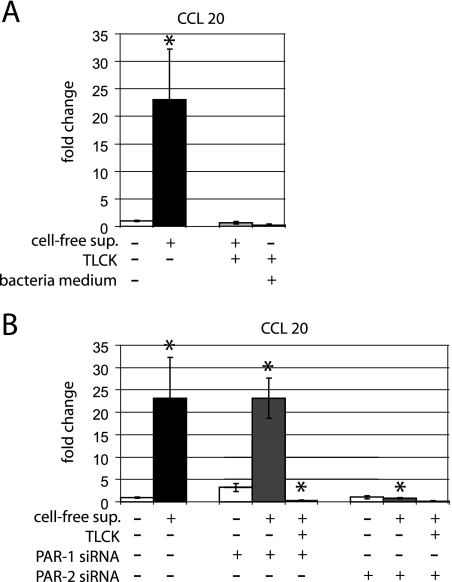
We next tested the effect of P. gingivalis supernatant on CCL20 mRNA expression because of the functional similarity of CCL20 and hBD-2 (55, 62). The gene expression of CCL20 was also significantly up-regulated (P = 0.05) in response to P. gingivalis cell-free supernatant (Fig. 5A). This effect was blocked by preincubation of supernatant with TLCK (P = 0.024). The blank bacterial medium and TLCK controls did not affect mRNA expression of hBD-2 (Fig. 5A).
FIG. 5.
CCL20 mRNA expression in response to P. gingivalis supernatant and effects of siRNA for PAR-1 and PAR-2 in gingival epithelial cells. (A) The mRNA expression of CCL20 was significantly up-regulated in response to the supernatant (P = 0.05). Stimulation of gingival epithelial cells with TLCK-pretreated supernatant and the control with complete bacterial media showed significantly lower CCL20 mRNA expression levels compared to stimulation with cell-free supernatant from P. gingivalis (P = 0.05 and P = 0.048, respectively). (B) The gene expression of CCL20 was significantly up-regulated in PAR-1 gene knockdown cells in response to the supernatant compared to that in untransfected unstimulated cells (P = 0.013). No difference between nontransfected and transfected cells was demonstrated in response to P. gingivalis supernatant (P = 0.976). This effect was completely abolished by TLCK-pretreatment (P = 0.006). In contrast, in PAR-2 gene knockdown cells, the mRNA expression of CCL20 showed significantly lower levels in response to the supernatant compared to that in untransfected stimulated cells (P = 0.05). The gene expression study was performed with triplicate samples derived from two to five different donors. *, significant difference (P ≤ 0.05). Error bars indicate standard deviations.
PAR-1 gene knockdown cells showed a significant up-regulation of CCL20 mRNA (P = 0.013) with bacterial supernatant compared to that in unstimulated control cells, and the effect was blocked by TLCK-preincubated supernatant (P = 0.006) (Fig. 5B). In contrast, PAR-2 gene knockdown cells showed no stimulation of CCL20 mRNA with P. gingivalis supernatant. Here the CCL20 gene expression was significantly lower than that in untransfected gingival cells treated with P. gingivalis cell-free supernatant (P = 0.05). The expression profile of CCL20 was comparable to that in unstimulated control cells (Fig. 5B). Treatment of control cells with the transfection agent HiPerFect showed no alteration of the CCL20 mRNA expression (data not shown). The magnitude of CCL20 gene expression varied in cells from different donors, but the overall pattern of effects of P. gingivalis supernatant and siRNA gene knockdown was consistent.
Purified RgpB regulates hBD-2 and CCL20 mRNA expression via PAR-2.
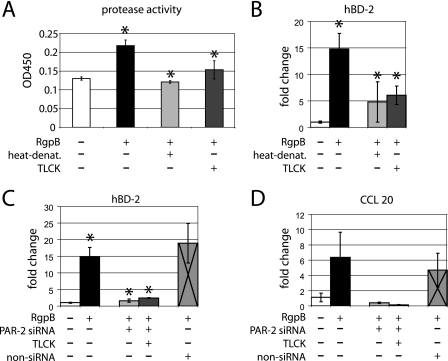
To confirm that proteases in the supernatant of P. gingivalis are responsible for the stimulation of hBD-2 and CCL20 mRNAs and to exclude other factors in the supernatant that may also have caused these effects, we next tested purified RgpB on gingival keratinocytes. For comparison, the proteolytic activity of RgpB was tested by the same multisubstrate assay method that was used for P. gingivalis cell-free supernatant. Compared to the control, RgpB showed proteolytic activity that was reduced by heat denaturation and TLCK treatment prior to incubation (P = 0.006 and 0.028, respectively) (Fig. 6A).
FIG. 6.
Purified RgpB activity and effects on hBD-2 and CCL20 mRNA expression evaluated by real-time PCR. (A) Protease activity (PDQ protease activity assay) of purified RgpB and TLCK-preincubated or heat-denaturated RgpB from P. gingivalis after 16 h. Compared to the assay control, purified RgpB showed significantly higher proteolytic activity after 16 h (P = 0.009). Purified RgpB showed significantly higher proteolytic activity than TLCK-preincubated or heat-denaturated RgpB (P = 0.028 and P = 0.006). Triplicate measurements were performed, and the values were normalized to blank control samples. (B) hBD-2 mRNA was significantly up-regulated in response to RgpB compared to that in unstimulated control cells (P = 0.016). This effect was abrogated by heat denaturation and TLCK pretreatment (P = 0.047 and 0.05, respectively). (C) In response to purified RgpB, the gene expression of hBD-2 was significantly lower in PAR-2 siRNA-transfected cells than in untransfected gingival epithelial cells (P = 0.024). The same observation was made for TLCK pretreatment of RgpB (P = 0.014). Cells transfected with scrambled nonsilencing RNA showed a significantly higher mRNA expression of hBD-2 in response to purified RgpB than unstimulated control cells (P = 0.037). (D) CCL20 mRNA was up-regulated in response to RgpB compared to that in unstimulated control cells. While not significant statistically, this effect was abrogated in PAR-2 gene knockdown cells (P = 0.095). The same observation was shown with TLCK pretreatment of RgpB (P = 0.084). Cells transfected with scrambled nonsilencing RNA showed a higher but not statistically significant mRNA expression of hBD-2 in response to purified RgpB than unstimulated control cells (P = 0.072). The gene expression study was performed in triplicate samples derived from two to five different donors. *, significant difference (P ≤ 0.05). Error bars indicate standard deviations.
The gene expression of hBD-2 was significantly up-regulated in response to the purified enzyme RgpB compared to that in unstimulated epithelial cells (P = 0.016) (Fig. 6B). This effect was diminished by heat denaturation and TLCK preincubation of RgpB (P = 0.05 and 0.047, respectively) (Fig. 6B). hBD-2 gene expression in response to RgpB in PAR-2 gene knockdown cells was significantly lower than in untransfected cells (P = 0.024) (Fig. 6C). TLCK preincubation of RgpB prior to stimulation of PAR-2 gene knockdown cells did not alter hBD-2 mRNA expression in gingival epithelial cells. Here the gene expression of hBD-2 was comparable to that in control cells and significantly lower than that in untransfected but RgpB-stimulated cells (P = 0.014) (Fig. 6C).
While not significant statistically, the CCL20 mRNA showed the same expression pattern as hBD-2 in response to RgpB, with up-regulation that was blocked in PAR-2 gene knockdown cells (P = 0.095) or in cells stimulated with TLCK-pretreated enzyme (P = 0.084) (Fig. 6D). The stimulation with RgpB resulted in up-regulation of hBD-2 and CCL20 mRNAs in cells transfected by scrambled nonsilencing control RNA and in untransfected cells (Fig. 6C and D).
DISCUSSION
Gingival epithelial cells respond to oral bacteria with up-regulation of the antimicrobial peptide hBD-2, an apparent protective response in the oral environment. Multiple signaling pathways are involved in this response to commensal bacteria and cell wall components (34, 35). However, cell wall preparations of P. gingivalis used previously failed to up-regulate hBD-2 (35), leading to the investigation of other pathways, such as PARs (4). In the present study, we report that proteolytic activity in P. gingivalis cell-free supernatant and purified Arg-gingipain specifically up-regulate the gene expression of hBD-2 and CCL20 and that this occurs via the action of PAR-2 but not via PAR-1. Surprisingly, this study also demonstrated the ability of P. gingivalis proteases to regulate the gene expression of protease-activated receptors in gingival epithelial cells. Both the PAR-1 and PAR-2 genes were affected, but in opposite ways. PAR-1 mRNA expression was down-regulated, whereas PAR-2 was up-regulated. Both effects were completely abrogated by the use of the protease inhibitor TLCK. This result suggests that gingipains secreted by P. gingivalis are involved in innate immune responses via PARs and that this effect may be enhanced by altering the expression of PAR-1 and PAR-2 mRNA in epithelial cells.
Arg-gingipains secreted by P. gingivalis are considered major etiologic factors in the development of periodontitis. In addition, Rgps have been shown to play a crucial role in controlling the expression of virulence factors and processing both extracellular and cell surface proteins of P. gingivalis (30). Therefore, we aimed to specifically test P. gingivalis proteases on gingival epithelial cells by using the cell-free supernatant and purified protease. Both P. gingivalis supernatant and purified RgpB had a LPS content of ≥1 endotoxin units/ml (Pyrochrome Limulus amebocyte lysate assay; Cape Cod, East Falmouth, MA) (data not shown). Although LPS was detected in the supernatant and the purified protease, effects on the gene expression of hBD-2 and CCL20 could not attributed to LPS in these preparations because pretreatment with the protease inhibitor TLCK or purified RgpB heat denaturation blocked up-regulation. These results are consistent with our previous findings in which P. gingivalis mutants (whole cell) lacking gingipains were ineffective in inducing the gene expression of hBD-2 in comparison to wild-type P. gingivalis (4). In addition, purified LPS exhibits poor stimulatory effects on the gene expression of hBD-2 in skin and oral epithelial cells (29, 35, 54). Thus, the effects reported here on the gene expression of hBD-2 and CCL20 were most likely based on proteases rather than the presence of LPS.
Previous findings led us to suggest that the proteases secreted by P. gingivalis are involved in antimicrobial peptide regulation via PARs (4). PARs are involved in both inflammatory and anti-inflammatory responses of epithelia (5, 6, 40). Thus, it was of interest to determine whether P. gingivalis regulates the gene expression of hBD-2 via PAR-1, PAR-2, or both PAR-1 and PAR-2, the major PAR family members present in gingival epithelial cells (40, 59). To address the specific role of PAR-1 versus PAR-2 we used the RNA interference technique. RNA interference allows sequence-specific, posttranscriptional gene silencing using siRNA that is a sequence homolog to the target gene (11, 14, 56, 57). Gingival epithelial cells were efficiently transfected using siRNA targeting the sequences for PAR-1 or PAR-2. The present results show that the gene expression of hBD-2 and CCL20 was up-regulated in response to protease-containing supernatant from P. gingivalis and by purified RgpB via PAR-2, extending previous findings (4). The specific role of PAR-2 was shown by the nearly complete loss of the stimulatory effect in PAR-2 but not in PAR-1 gene knockdown cells. The findings were supported by controls using TLCK-treated samples. This study demonstrates the importance of the protease in the P. gingivalis cell-free supernatant rather than the presence of other possible virulence factors such as LPS. Thus, the gene expression of hBD-2 is mediated via proteases that signal via PAR-2 but not via PAR-1.
The antimicrobial peptide hBD-2 and the structurally related chemokine CCL20 both attract immature dendritic cells (2, 9, 55) and interact with chemokine receptor 6 (61). Here, we report for the first time that proteases secreted by P. gingivalis are responsible for the increased gene expression of CCL20 as well as hBD-2 via a PAR-2-mediated mechanism. CCL20 mRNA expression induced by P. gingivalis cell-free supernatant was completely abrogated in PAR-2 gene knockdown cells. These results suggest parallel responses in gene expression of hBD-2 and CCL20. These findings indicate that the epithelial innate immune response to P. gingivalis may also lead to a synergistic response and an amplified activation of dendritic cells by a parallel expression of both hBD-2 and CCL20 in the gingival epithelium.
There has been controversy in the literature over the role of PARs. In some tissues they are associated with inflammation (42), while in other tissues they seem to have an anti-inflammatory or protective effect (31, 32). The fact that hBD-2 gene expression is up-regulated via PAR-2 suggests a possible protective function promoted by PAR-2 in oral epithelial cells. This protective function may be facilitated by the up-regulation of PAR-2 and the down-regulation of PAR-1 gene expression in response to P. gingivalis proteases. The response to P. gingivalis suggests a cytoprotective epithelial autoregulatory mechanism that maintains or increases the level of PAR-2 which responds to the potential danger of the presence of P. gingivalis proteases via hBD-2 and CCL20 expression. Since PAR-1, -3, and -4 are activated by thrombin, whereas PAR-2 is activated by trypsin and proteases other than thrombin (26, 40, 58), the PAR-2 receptor might play a unique role among PARs that allows epithelial cells to recognize pathogenic bacteria that promote innate immune responses.
In conclusion, we report here that gingival epithelial cells utilize PAR-2 to mediate the gene expression of hBD-2 and CCL20 in response to proteases secreted by P. gingivalis. These findings show the pronounced ability of gingival epithelial cells to maintain their protective function by recognizing pathogenic bacteria and synthesizing antimicrobial peptides. This possible cytoprotective role mediated by PAR-2 will be explored in future studies aiming for a better understanding of the development and treatment of gingival inflammatory diseases.
Acknowledgments
We kindly thank Janet R. Kimball, Department of Oral Biology, University of Washington, for excellent technical assistance and critically reading the manuscript. We thank Beth Hacker and Theresa Oswald at the University of Washington Comprehensive Center for Oral Health Research for providing the gingival epithelial cells. We also thank Pam Braham for her help with the bacteria culture.
This work was funded by NIDCR grant R01DE16961.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 25 June 2007.
REFERENCES
- 1.Belton, C. M., K. T. Izutsu, P. C. Goodwin, Y. Park, and R. J. Lamont. 1999. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell. Microbiol. 1:215-223. [DOI] [PubMed] [Google Scholar]
- 2.Biragyn, A., P. A. Ruffini, C. A. Leifer, E. Klyushnenkova, A. Shakhov, O. Chertov, A. K. Shirakawa, J. M. Farber, D. M. Segal, J. J. Oppenheim, and L. W. Kwak. 2002. Toll-like receptor 4-dependent activation of dendritic cells by beta-defensin 2. Science 298:1025-1029. [DOI] [PubMed] [Google Scholar]
- 3.Chung, W. O., and B. A. Dale. 2004. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect. Immun. 72:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, W. O., S. R. Hansen, D. Rao, and B. A. Dale. 2004. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J. Immunol. 173:5165-5170. [DOI] [PubMed] [Google Scholar]
- 5.Cottrell, G. S., S. Amadesi, F. Schmidlin, and N. Bunnett. 2003. Protease-activated receptor 2: activation, signalling and function. Biochem. Soc. Trans. 31:1191-1197. [DOI] [PubMed] [Google Scholar]
- 6.Coughlin, S. R., and E. Camerer. 2003. PARticipation in inflammation. J. Clin. Investig. 111:25-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale, B. A., J. R. Kimball, S. Krisanaprakornkit, F. Roberts, M. Robinovitch, R. O'Neal, E. V. Valore, T. Ganz, G. M. Anderson, and A. Weinberg. 2001. Localized antimicrobial peptide expression in human gingiva. J. Periodontal Res. 36:285-294. [DOI] [PubMed] [Google Scholar]
- 8.Dommisch, H., Y. Acil, A. Dunsche, J. Winter, and S. Jepsen. 2005. Differential gene expression of human beta-defensins (hBD-1, -2, -3) in inflammatory gingival diseases. Oral Microbiol. Immunol. 20:186-190. [DOI] [PubMed] [Google Scholar]
- 9.Duits, L. A., B. Ravensbergen, M. Rademaker, P. S. Hiemstra, and P. H. Nibbering. 2002. Expression of beta-defensin 1 and 2 mRNA by human monocytes, macrophages and dendritic cells. Immunology 106:517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunsche, A., Y. Acil, R. Siebert, J. Harder, J. M. Schroder, and S. Jepsen. 2001. Expression profile of human defensins and antimicrobial proteins in oral tissues. J. Oral Pathol. Med. 30:154-158. [DOI] [PubMed] [Google Scholar]
- 11.Fire, A. 1999. RNA-triggered gene silencing. Trends Genet. 15:358-363. [DOI] [PubMed] [Google Scholar]
- 12.Fujimura, S., K. Hirai, Y. Shibata, K. Nakayama, and T. Nakamura. 1998. Comparative properties of envelope-associated arginine-gingipains and lysine-gingipain of Porphyromonas gingivalis. FEMS Microbiol. Lett. 163:173-179. [DOI] [PubMed] [Google Scholar]
- 13.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 14.Hammond, S. M., A. A. Caudy, and G. J. Hannon. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2:110-119. [DOI] [PubMed] [Google Scholar]
- 15.Hancock, R. E. 1997. Peptide antibiotics. Lancet 349:418-422. [DOI] [PubMed] [Google Scholar]
- 16.Harder, J., J. Bartels, E. Christophers, and J. M. Schroder. 1997. A peptide antibiotic from human skin. Nature 387:861. [DOI] [PubMed] [Google Scholar]
- 17.Harder, J., U. Meyer-Hoffert, L. M. Teran, L. Schwichtenberg, J. Bartels, S. Maune, and J. M. Schroder. 2000. Mucoid Pseudomonas aeruginosa, TNF-alpha, and IL-1beta, but not IL-6, induce human beta-defensin-2 in respiratory epithelia. Am. J. Respir. Cell Mol. Biol. 22:714-721. [DOI] [PubMed] [Google Scholar]
- 18.Harder, J., U. Meyer-Hoffert, K. Wehkamp, L. Schwichtenberg, and J. M. Schroder. 2004. Differential gene induction of human beta-defensins (hBD-1, -2, -3, and -4) in keratinocytes is inhibited by retinoic acid. J. Investig. Dermatol. 123:522-529. [DOI] [PubMed] [Google Scholar]
- 19.Hasebe, A., A. Yoshimura, T. Into, H. Kataoka, S. Tanaka, S. Arakawa, H. Ishikura, D. T. Golenbock, T. Sugaya, N. Tsuchida, M. Kawanami, Y. Hara, and K. Shibata. 2004. Biological activities of Bacteroides forsythus lipoproteins and their possible pathological roles in periodontal disease. Infection Immun. 72:1318-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hintermann, E., S. K. Haake, U. Christen, A. Sharabi, and V. Quaranta. 2002. Discrete proteolysis of focal contact and adherens junction components in Porphyromonas gingivalis-infected oral keratinocytes: a strategy for cell adhesion and migration disabling. Infect. Immun. 70:5846-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano, K., T. Yufu, M. Hirano, J. Nishimura, and H. Kanaide. 2005. Physiology and pathophysiology of proteinase-activated receptors (PARs): regulation of the expression of PARs. J. Pharmacol. Sci. 97:31-37. [DOI] [PubMed] [Google Scholar]
- 22.Holt, S. C., and J. L. Ebersole. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000 38:72-122. [DOI] [PubMed] [Google Scholar]
- 23.Holt, S. C., L. Kesavalu, S. Walker, and C. A. Genco. 1999. Virulence factors of Porphyromonas gingivalis. Periodontology 2000 20:168-238. [DOI] [PubMed] [Google Scholar]
- 24.Holzhausen, M., L. C. Spolidorio, R. P. Ellen, M. C. Jobin, M. Steinhoff, P. Andrade-Gordon, and N. Vergnolle. 2006. Protease-activated receptor-2 activation: a major role in the pathogenesis of Porphyromonas gingivalis infection. Am. J. Pathol. 168:1189-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holzhausen, M., L. C. Spolidorio, and N. Vergnolle. 2005. Proteinase-activated receptor-2 (PAR2) agonist causes periodontitis in rats. J. Dent. Res. 84:154-159. [DOI] [PubMed] [Google Scholar]
- 26.Holzhausen, M., L. C. Spolidorio, and N. Vergnolle. 2005. Role of protease-activated receptor-2 in inflammation, and its possible implications as a putative mediator of periodontitis. Mem. Inst. Oswaldo Cruz 100(Suppl. 1):177-180. [DOI] [PubMed] [Google Scholar]
- 27.Hoover, D. M., C. Boulegue, D. Yang, J. J. Oppenheim, K. Tucker, W. Lu, and J. Lubkowski. 2002. The structure of human macrophage inflammatory protein-3alpha /CCL20. Linking antimicrobial and CC chemokine receptor-6-binding activities with human beta-defensins. J. Biol. Chem. 277:37647-37654. [DOI] [PubMed] [Google Scholar]
- 28.Joly, S., C. Maze, P. B. McCray, Jr., and J. M. Guthmiller. 2004. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J. Clin. Microbiol. 42:1024-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joly, S., C. C. Organ, G. K. Johnson, P. B. McCray, Jr., and J. M. Guthmiller. 2005. Correlation between beta-defensin expression and induction profiles in gingival keratinocytes. Mol. Immunol. 42:1073-1084. [DOI] [PubMed] [Google Scholar]
- 30.Kadowaki, T., K. Nakayama, F. Yoshimura, K. Okamoto, N. Abe, and K. Yamamoto. 1998. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 273:29072-29076. [DOI] [PubMed] [Google Scholar]
- 31.Kawabata, A., H. Nishikawa, H. Saitoh, Y. Nakaya, K. Hiramatsu, S. Kubo, M. Nishida, N. Kawao, R. Kuroda, F. Sekiguchi, M. Kinoshita, K. Kakehi, N. Arizono, H. Yamagishi, and K. Kawai. 2004. A protective role of protease-activated receptor 1 in rat gastric mucosa. Gastroenterology 126:208-219. [DOI] [PubMed] [Google Scholar]
- 32.Kawabata, A., Y. Oono, D. Yonezawa, K. Hiramatsu, N. Inoi, F. Sekiguchi, M. Honjo, M. Hirofuchi, T. Kanke, and H. Ishiwata. 2005. 2-Furoyl-LIGRL-NH2, a potent agonist for proteinase-activated receptor-2, as a gastric mucosal cytoprotective agent in mice. Br. J. Pharmacol. 144:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krisanaprakornkit, S., D. Jotikasthira, and B. A. Dale. 2003. Intracellular calcium in signaling human beta-defensin-2 expression in oral epithelial cells. J. Dent. Res. 82:877-882. [DOI] [PubMed] [Google Scholar]
- 34.Krisanaprakornkit, S., J. R. Kimball, and B. A. Dale. 2002. Regulation of human beta-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathways, but not the NF-kappaB transcription factor family. J. Immunol. 168:316-324. [DOI] [PubMed] [Google Scholar]
- 35.Krisanaprakornkit, S., J. R. Kimball, A. Weinberg, R. P. Darveau, B. W. Bainbridge, and B. A. Dale. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehrer, R. I. 2004. Primate defensins. Nat. Rev. Microbiol. 2:727-738. [DOI] [PubMed] [Google Scholar]
- 39.Lourbakos, A., C. Chinni, P. Thompson, J. Potempa, J. Travis, E. J. Mackie, and R. N. Pike. 1998. Cleavage and activation of proteinase-activated receptor-2 on human neutrophils by gingipain-R from Porphyromonas gingivalis. FEBS Lett. 435:45-48. [DOI] [PubMed] [Google Scholar]
- 40.Lourbakos, A., J. Potempa, J. Travis, M. R. D'Andrea, P. Andrade-Gordon, R. Santulli, E. J. Mackie, and R. N. Pike. 2001. Arginine-specific protease from Porphyromonas gingivalis activates protease-activated receptors on human oral epithelial cells and induces interleukin-6 secretion. Infect. Immun. 69:5121-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lourbakos, A., Y. P. Yuan, A. L. Jenkins, J. Travis, P. Andrade-Gordon, R. Santulli, J. Potempa, and R. N. Pike. 2001. Activation of protease-activated receptors by gingipains from Porphyromonas gingivalis leads to platelet aggregation: a new trait in microbial pathogenicity. Blood 97:3790-3797. [DOI] [PubMed] [Google Scholar]
- 42.Lu, C., F. D. Zhao, X. B. Li, and L. H. Yin. 2005. Up regulation of interleukin-8 expressions induced by mast cell tryptase via protease activated receptor-2 in endothelial cell line. Chin. Med. J. 118:1900-1906. [PubMed] [Google Scholar]
- 43.MacNaughton, W. K. 2005. Epithelial effects of proteinase-activated receptors in the gastrointestinal tract. Mem. Inst. Oswaldo Cruz 100(Suppl. 1):211-215. [DOI] [PubMed] [Google Scholar]
- 44.Mathews, M., H. P. Jia, J. M. Guthmiller, G. Losh, S. Graham, G. K. Johnson, B. F. Tack, and P. B. McCray, Jr. 1999. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect. Immun. 67:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicolas, P., and A. Mor. 1995. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu. Rev. Microbiol. 49:277-304. [DOI] [PubMed] [Google Scholar]
- 46.O'Neil, D. A., E. M. Porter, D. Elewaut, G. M. Anderson, L. Eckmann, T. Ganz, and M. F. Kagnoff. 1999. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 163:6718-6724. [PubMed] [Google Scholar]
- 47.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Potempa, J., R. Pike, and J. Travis. 1995. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect. Immun. 63:1176-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potempa, J., and J. Travis. 1996. Porphyromonas gingivalis proteinases in periodontitis, a review. Acta Biochim. Pol. 43:455-465. [PubMed] [Google Scholar]
- 50.Rangarajan, M., S. J. Smith, S. U, and M. A. Curtis. 1997. Biochemical characterization of the arginine-specific proteases of Porphyromonas gingivalis W50 suggests a common precursor. Biochem. J. 323:701-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren, L., W. K. Leung, R. P. Darveau, and L. Jin. 2005. The expression profile of lipopolysaccharide-binding protein, membrane-bound CD14, and Toll-like receptors 2 and 4 in chronic periodontitis. J. Periodontol. 76:1950-1959. [DOI] [PubMed] [Google Scholar]
- 52.Rudney, J. D., R. Chen, and G. J. Sedgewick. 2001. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect. Immun. 69:2700-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandros, J., P. N. Papapanou, U. Nannmark, and G. Dahlen. 1994. Porphyromonas gingivalis invades human pocket epithelium in vitro. J. Periodontal Res. 29:62-69. [DOI] [PubMed] [Google Scholar]
- 54.Schauber, J., R. A. Dorschner, K. Yamasaki, B. Brouha, and R. L. Gallo. 2006. Control of the innate epithelial antimicrobial response is cell-type specific and dependent on relevant microenvironmental stimuli. Immunology 118:509-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schutyser, E., S. Struyf, and J. Van Damme. 2003. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 14:409-426. [DOI] [PubMed] [Google Scholar]
- 56.Sharp, P. A. 2001. RNA interference—2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 57.Tuschl, T. 2001. RNA interference and small interfering RNAs. Chembiochemistry 2:239-245. [DOI] [PubMed] [Google Scholar]
- 58.Uehara, A., K. Muramoto, H. Takada, and S. Sugawara. 2003. Neutrophil serine proteinases activate human nonepithelial cells to produce inflammatory cytokines through protease-activated receptor 2. J. Immunol. 170:5690-5696. [DOI] [PubMed] [Google Scholar]
- 59.Uehara, A., S. Sugawara, K. Muramoto, and H. Takada. 2002. Activation of human oral epithelial cells by neutrophil proteinase 3 through protease-activated receptor-2. J. Immunol. 169:4594-4603. [DOI] [PubMed] [Google Scholar]
- 60.Vergnolle, N. 2005. Clinical relevance of proteinase activated receptors (pars) in the gut. Gut 54:867-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, D., Q. Chen, O. Chertov, and J. J. Oppenheim. 2000. Human neutrophil defensins selectively chemoattract naive T and immature dendritic cells. J. Leukoc. Biol. 68:9-14. [PubMed] [Google Scholar]
- 62.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schroder, J. M. Wang, O. M. Howard, and J. J. Oppenheim. 1999. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]