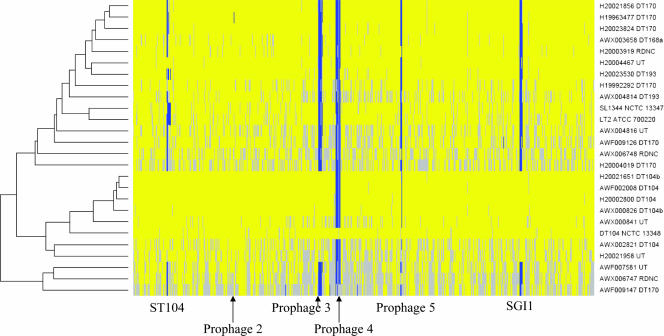
FIG. 1.
Analysis of the microarray data by GACK. Test/reference ratios of the 24 S. enterica serovar Typhimurium isolates analyzed by microarray were assessed for the presence/absence of genes by using GACK software. The input data set was restricted to the 4,167 chromosomal features (SDT loci) expected to be present in one or more of the isolates. The heat map is plotted in the physical order of the SDT loci according to the order of the loci in the S. enterica serovar Typhimurium DT104 (NCTC 13348) genome. The presence of a gene in the test strain that is also present in the control strain (S. enterica serovar Typhimurium DT104 [NCTC 13348]) is shown in yellow, the absence or divergence of the gene from its presence in the control strain is shown in blue, and genes for which their presence was uncertain are in gray. The positions of prophage 1/ST104 and prophages 2 to 5 as well as SGI1 are indicated.