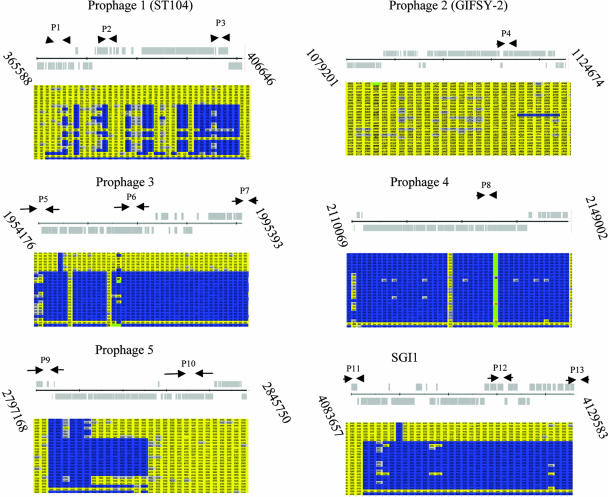
FIG. 2.
Schematic diagram of prophages 1 to 5 and SGI1, showing oligonucleotide target sites. The gray schematic diagrams represent the predicted open reading frames in the annotated genome of S. enterica serovar Typhimurium DT104 (NCTC 13348). The arrows show the location of the primer pairs P1 to P13, as listed in Table 3. The corresponding microarray results, processed by the gene-calling program GACK, are shown underneath each prophage or under SGI1. The columns represent the features harbored in each prophage or the genomic island. Yellow represents features that are present in the test strain as well as the control strain (S. enterica serovar Typhimurium DT104 [NCTC 13348]), blue represents genes that are in the control strain but are absent in the test strain, and gray indicates that the presence of the gene in the test strain is uncertain. The rows represent each S. enterica serovar Typhimurium isolate, in the following order: row 1, AWF002008; 2, AWX002821; 3, H20002800; 4, H20021651; 5, AWX000826; 6, AWX000841; 7, H20021958; 8, H20004019; 9, AWF009126; 10, AWF009147; 11, H20021856; 12, AWX003658; 13, AWX004816; 14, AWF007581; 15, H19963477; 16, H19992292; 17, AWX004814; 18, H20004467; 19, H20023824; 20 AWX006747; 21, AWX006748; 22, H20003919; 23, H20023530; 24, DT104 (NCTC 13348); 25, LT2 (ATCC 700220).