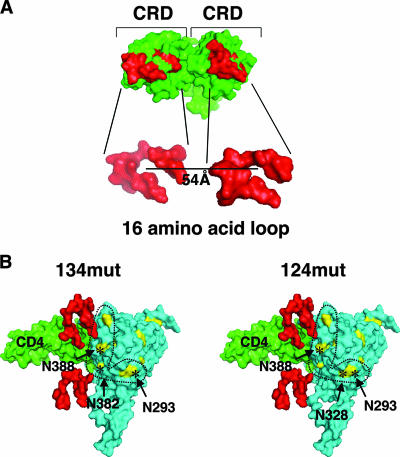
FIG. 6.
Pairs of N-glycan sites in gp120 can fit the spatial requirements of the CRDs in the DC-SIGN dimer. (A) The crystal structure of the DC-SIGN CRD as a dimer is shown as a space-filling model. The 16-amino-acid loop that forms the carbohydrate binding region of each CRD of DC-SIGN is highlighted in red (Protein Data Bank identification no., 1XAR). (B) A scaled model of the dimeric CRDs of DC-SIGN is positioned next to a space-filling model of the crystal structure of the CD4-gp120 complex (the V3 loop is included; Protein Data Bank identification no., 2B4C). For clarity, only the 16-amino-acid loop that forms the carbohydrate binding region is shown (red). CD4 is in green, and gp120 is in blue. High-mannose N-glycan sites are shown in yellow, and the specific sites mutated in the 134mut and 124mut envelopes are indicated by asterisks in the respective models. The five N-glycan sites that make up the potential 2G12 epitope are those within the dotted lines. The scaled modeling of this composite structure is described in Materials and Methods.