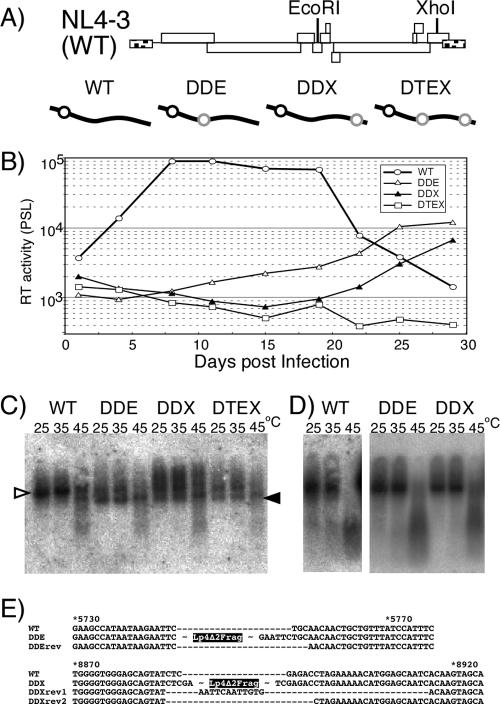
FIG. 5.
Replication assay of mutants carrying a monomeric genome. (A) Schematic diagrams of replication-competent form mutants. The positions of restriction enzyme sites on the viral genome used for insertion are shown in the upper part of the panel. Diagrams of the mutants are shown in the lower part of the panel. Symbols are the same as those described for Fig. 3. (B) Growth kinetics of viruses. Values are representative of the results of at least three independent experiments. Viruses were prepared by transfection of 293T cells with pNL4-3 (WT) or its derivative mutants (pDDEE+ [DDE], pDDXE+ [DDX], and pDTEXE+ [DTE]). At 48 h posttransfection, culture supernatants of transfected 293T cells were harvested, and equal quantities of CA-p24 were inoculated into MT-4 cells. The supernatants of the cells were harvested every 3 or 4 days. Ten microliters of each cell supernatant was subjected to exogenous RT assay and quantitated by PhosphorImager analysis. PSL, Photostimulierte Lumineszenz. (C) Virion RNA profiles produced from transfected 293T cells and visualized by native Northern blotting analysis. Open and solid arrowheads denote positions of dimers and monomers, respectively. (D) Virion RNA profiles produced from MT-4 cells. Viruses were harvested at their growth kinetic peak point (wild type, 10 days postinfection; DDE and DDX, 28 days postinfection). (E) The nature of reversions. The sequences in the vicinity of the fragment-inserted sites are shown. The names of revertant sequences include “rev.” The positions of Lp4Δ2 fragment insertion of DDE and DDX are indicated. The numbers above the sequences represent nucleotide positions of pNL4-3 (WT).