Abstract
Over the past 2 decades, significant effort has been dedicated to the development of adeno-associated virus (AAV) as a vector for human gene therapy. However, understanding of the virus with respect to the functional domains of the capsid remains incomplete. In this study, the goal was to further examine the role of the unique Vp1 N terminus, the N terminus plus the recently identified nuclear localization signal (NLS) (J. C. Grieger, S. Snowdy, and R. J. Samulski, J. Virol 80:5199-5210, 2006), and the virion pore at the fivefold axis in infection. We generated two Vp1 fusion proteins (Vp1 and Vp1NLS) linked to the 8-kDa chemokine domain of rat fractalkine (FKN) for the purpose of surface exposure upon assembly of the virion, as previously described (K. H. Warrington, Jr., O. S. Gorbatyuk, J. K. Harrison, S. R. Opie, S. Zolotukhin, and N. Muzyczka, J. Virol 78:6595-6609, 2004). The unique Vp1 N termini were found to be exposed on the surfaces of these capsids and maintained their phospholipase A2 (PLA2) activity, as determined by native dot blot Western and PLA2 assays, respectively. Incorporation of the fusions into AAV type 2 capsids lacking a wild-type Vp1, i.e., Vp2/Vp3 and Vp3 capsid only, increased infectivity by 3- to 5-fold (Vp1FKN) and 10- to 100-fold (Vp1NLSFKN), respectively. However, the surface-exposed fusions did not restore infectivity to AAV virions containing mutations at a conserved leucine (Leu336Ala, Leu336Cys, or Leu336Trp) located at the base of the fivefold pore. EM analyses suggest that Leu336 may play a role in global structural changes to the virion directly impacting downstream conformational changes essential for infectivity and not only have local effects within the pore, as previously suggested.
Adeno-associated virus (AAV) is a member of the parvovirus family. It is a single-stranded DNA virus with a genome of ∼4.7 kb that is dependent upon coinfection with helper viruses, such as adenovirus or herpesvirus, for efficient reproduction (5, 7). This small genome encodes four nonstructural proteins (Rep78, -68, -54, and -48) and three viral capsid proteins, Vp1, Vp2, and Vp3 (29, 36). The Rep proteins are multifunctional and play roles in almost every aspect of the life cycle of AAV, including replication, transcription, integration, and packaging of the genome into the preformed empty capsid (10, 23). Vp1, Vp2, and Vp3 are incorporated into capsids at a predicted ratio of 1:1:8 and have overlapping sequences, differing only at their N termini. Vp2 is 137 amino acids shorter than Vp1 and is the product of an alternative start codon, while Vp3 is 65 residues shorter than Vp2.
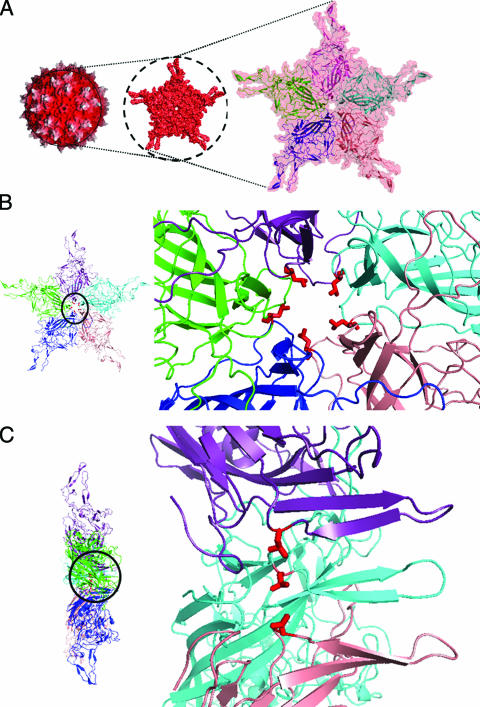
The structure of AAV type 2 (AAV2) has been determined at 3-Å resolution by X-ray crystallography. In addition, the capsid structures of other parvoviruses have been resolved by X-ray crystallography or cryoelectron microscopy (cryo-EM) and image reconstruction (1-3, 25, 26, 31, 34, 40, 42, 45, 48, 49). The parvovirus virion is composed of 60 subunits of the overlapping Vp (∼530 C-terminal amino acids) region arranged with T = 1 icosahedral symmetry (25, 48). Common features are a depression at the twofold axis of symmetry, three protrusions at or surrounding the threefold axes, and 12 narrow pores at the fivefold axes. The overlapping subunit structure is comprised of an eight-stranded β-barrel motif containing two antiparallel β-sheets (48). Long loop insertions between the strands comprise about 60% of the structure. These interstrand loops contain small regions of β-structure and a conserved helix that forms the wall of the depressions at the twofold axes. Two small stretches of strand structure form a β-ribbon in each subunit, which cluster at the icosahedral fivefold axis of the capsid to create the cylindrical pores that connect the inside of the capsid to the outside. The 12 narrow pores at the fivefold axes are one of the most striking structural features of AAV2 capsids (Fig. 1). The pores protrude from the surrounding capsid surface, surrounded by a depression that is lined by a loop between the βH and βI strands of the β-barrel. It is thought that the β-ribbons are dynamic structures, since the pore increases in diameter from the inside to the outside of the capsid. The β-ribbons are made up of residues 322 to 338 of the capsid sequence, including a highly conserved Leu336, and the residue type is found in all known parvoviruses. This residue surrounds the inside base of the channel, with side chains constricting the channel to ∼8.7 Å in diameter (Fig. 1B and C).
FIG. 1.
AAV2 crystal structure. (A) Surface topology of the whole capsid, viewed down the icosahedral fivefold symmetry axis, that contains a pore (left) bounded by five symmetry-related Vp subunits shown in salmon, blue, green, purple, and cyan ribbons in the close-up view on the right. The depth-cued view (left) shows the tops of the three protrusions surrounding the icosahedral threefold axes in white and the rest of the capsid in red. (B) Top and (C) side views of the icosahedral fivefold-symmetry-related Vps (in the same colors as in panel A) with a close-up of the pore on the right. Leu336, located at the inside base of the pore, is highlighted in red in panels B and C.
The unique N termini of Vp1 and Vp2, proposed to be localized on the inside of the capsids, have not been visualized in any of the parvovirus crystal structures, although cryo-EM and image reconstruction studies of AAV2 interpreted “globules” observed under the twofold axis as structural candidates for these regions (24, 25). In the crystal structure of mature MVM virions, weak density within the fivefold pores could be modeled as a glycine-rich sequence within the N terminus of Vp1 to Vp3 of minute virus of mice (MVM) (3). The crystal structure of canine parvovirus (CPV) virions supports the hypothesis that the weak density inside the fivefold pore corresponds to N-terminal peptide sequences (49). There is biochemical and mutational evidence that the N-terminus of Vp1 and Vp2 in some parvoviruses can be externalized onto the capsid surface via the pores at the fivefold axes while the capsid remains intact. In parvoviruses, such as MVM and CPV, that undergo maturation cleavage of Vp2 to produce Vp3 following genomic-DNA packaging, it is thought that this exposure allows the processing (13, 44). It is also important to note that the unique N-terminal region of Vp1 of CPV became exposed in vivo within endosomal/lysosomal vesicles (35, 37). In vitro exposure of Vp1 N termini has been achieved by treatment of capsids with heat or urea (8, 13, 41). However, in empty CPV, MVM, and AAV2 capsids, the N-terminal regions do not become accessible to antibodies while the capsid remains intact (13, 25, 41), suggesting that the viral genome aids in the transition required for surface exposure.
The early steps of AAV2 infection begin with attachment to heparan sulfate proteoglycan and to a variety of cell surface receptors, such as FGFR, αVβ5 integrin, α5β1 integrin, and hepatocyte growth factor receptor (c-Met) (4, 14, 22, 32, 38, 39), followed by clathrin dependent endocytosis. Based on a number of studies, it has been proposed that AAV requires endosomal acidification to escape from the late endosome and traffic to the nucleus (16, 20, 50, 51). These studies also suggest that prior to escaping the endosome, the virion must undergo conformational changes leading to the exposure of the unique N terminus of Vp1 and the N terminus of Vp2 required for endososmal escape and nuclear entry (6, 9, 33, 35). The unique N terminus of Vp1 contains a conserved phospholipase A2 (PLA2) domain (8, 15, 18, 28, 52) that is essential for infectivity and is thought to be required for endosomal escape. A suggestion that the N terminus of Vp1 are located within the virion and become surface exposed naturally within the cell via an induced conformational change is consistent with the observation that while intact capsids do not have PLA2 activity, heat or acidic-pH treatment of virions elicits this function (37, 52). Basic amino acid clusters, which are thought to control the nuclear import of incoming virions, are also contained within the unique Vp1 N terminus (27, 41) and the N-terminal amino acid stretch (166PARKRLN172) contained within an overlapping region of Vp1 and Vp2 (19, 35). With increasing knowledge of capsid motifs involved in AAV infection, it is currently unknown if the virions that have escaped the endosome successfully infect the host cell, because it has been previously shown that nuclear entry of intact AAV virions is very inefficient, suggesting that uncoating may occur before or during nuclear entry (30, 47).
In addition, the pore at the fivefold axis of the parvovirus capsid has been postulated as the site for genome import into the preformed capsid, genome export for uncoating, and Vp1/Vp2 N-terminus exposure during trafficking (6, 35). The purpose of this study was to further explore the role of the fivefold pores in the life cycle of AAV2. We decided to take a unique approach to determine the significance of the pore in the life cycle of AAV2 by generating Vp1 protein fusion constructs that would place the unique N terminus of Vp1 on the outside of the virus, similar to the autonomous parvovirus B19. A previous study of AAV2 capsids generated with the 8-kDa chemokine domain of fractalkine (FKN) fused to the end of the Vp2 protein had shown that the fusion protein was displayed on the capsid surface (6). We rationalized that a construct made from a Vp2AFKN fusion to which the unique N-terminus of Vp1 was fused should also display the latter protein on the capsid surface. Our approach was to test the ability of such a fusion construct, carrying an active PLA2 domain with and without a nuclear localization signal (NLS) exposed on the capsid surface upon assembly, to enhance/restore the infectivity of Vp2/Vp3 and Vp3-only virions. In addition, we tested the abilities of these fusion constructs to rescue viruses with mutations at the conserved Leu336 residues positioned at the base of the fivefold axis, for which prevention of Vp1 N-terminus exposure through the fivefold pore had been reported to be detrimental to infection (6). The Vp1 fusion proteins incorporated into the AAV capsid were surface exposed and functional based on native dot blot Western blotting and a PLA2 assay. Infectivity was enhanced/regained for Vp2/Vp3 and Vp3-only virions when Vp1FKN and Vp1NLSFKN were incorporated into the virion. However, the Vp1 fusion proteins did not rescue/enhance infectivity for the Leu336 mutants. In addition, the Leu336 mutant virions appeared to have altered permeability to transmission EM-negative stain compared to virions assembled from wild-type (wt) viral capsid proteins. Taken together, our data support the conclusions that (i) it is possible to generate infectious AAV virions that surface display their Vp1 N termini upon assembly, as has been reported for B19, and (ii) mutations of the conserved Leu336 cannot be rescued by surface-exposed Vp1 N termini, suggesting that Leu336 mutants may lead to a global conformational change in virions or inhibit an essential structural change that prevents successful infection.
MATERIALS AND METHODS
Cells and viruses.
HeLa and 293 cells were maintained at 37°C in a 5% CO2 atmosphere in Dulbecco's modified Eagle's medium (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum and penicillin-streptomycin. Adenovirus dl309 has been described previously (21).
Plasmids.
The AAV2 helper plasmid (pXR2) was used throughout this study as a starting reagent to generate the fivefold pore mutant vectors. The following oligonucleotides were designed and utilized in the QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene) according to protocol to mutate the conserved leucine residue 336 to cysteine, tryptophan, and alanine (lowercase type represents the codon sequence in each primer sequence): AAV2-Leu336Cys (AAV2-cys), ACG ACG ATT GCC AAT AAC tgt ACC AGC ACG GTT CAG GTG; AAV2-Leu336Trp (AAV2-trp), ACG ACG ATT GCC AAT AAC tgg ACC AGC ACG GTT CAG GTG; and AAV2-Leu336Ala (AAV2-ala), ACG ACG ATT GCC AAT AAC gct ACC AGC ACG GTT CAG GTG. The Vp2/Vp3 and Vp3-only plasmids were also generated using pXR2 via mutagenesis of the Vp1 and the Vp1/Vp2 start codons, respectively, as previously published (43). The Vp2AFKN and pXR2 constructs were used to generate both Vp1 fusion constructs. Vp1FKN was generated by mutating the Vp1 start site of Vp2AFKN back to ATG (lowercase), ATT TAA ATC AGG Tat gGC TGC CGA TGG TTA TCT TCC, and Vp2A ATG start codon to leucine to prevent expression of Vp2AFKN.
Western blotting.
Samples from the cell homogenate and purified vector were loaded onto NuPage 10% Bis-Tris gels and run using 1× NuPage MOPS (morpholinepropanesulfonic acid) buffer. The XCell SureLock Mini Cell (Invitrogen) was used for electrophoresis. The protein was then transferred to a Hybond ECL membrane utilizing the XCell II Blot module (Invitrogen) for wet transfer according to the manufacturer's protocol for Bis-Tris gels. Each membrane was blocked for 1 h at room temperature using 1× Tris-buffered saline (TBS), pH 7.5, 0.1% Tween, and 10% nonfat dry milk (NFDM). The primary B1 antibody was then diluted 1:10 in 1× TBS, pH 7.5, 0.1% Tween, and 2% NFDM and incubated on the membrane for 1 h at room temperature or overnight at 4°C. Excess primary antibody was then washed from the membrane using 1× TBS, pH 7.5, 0.1% Tween four times for 5 min each time. Secondary antibody was then diluted 1:10,000 in 1× TBS, pH 7.5, 0.1% Tween, and 2% NFDM and incubated on the membrane for 1 h at room temperature. Excess secondary antibody was washed from the membrane as previously described. SuperSignal West Femto Maximum Sensitivity Substrate (Pierce) was added to each membrane according to the manufacturer's protocol. Each membrane was then exposed to Kodak BioMax MR film.
Production of rAAV.
To produce the recombinant AAV2 (rAAV2) vectors utilized in this study, the triple-transfection method was carried out utilizing polyethylenimine (linear molecular weight, ∼ 25,000). Twelve micrograms of XX680 (adenovirus helper plasmid), 10 μg of pXR2 or mutant helper plasmids, and 6 μg of CMV-luciferase plasmid were aliquotted into 1.5-ml microcentrifuge tubes (1 tube per 15-cm plate of 293 cells). Five hundred microliters of serum-free Dulbecco's modified Eagle's medium was then added to each tube, followed by the addition of 110 μl of 1-mg/ml polyethylenimine. Each transfection mixture was vortexed briefly and incubated at room temperature for 5 to 10 min. The transfection mixture was added directly to 15- cm plates of 293 cells. Five 15-cm plates were used to generate each virus. The cells were harvested 48 h posttransfection by scraping the cells from the plate and then pelleted by low-speed centrifugation (1,500 rpm). The cells were lysed, and the cell homogenate was loaded onto iodixanol step gradients as described previously (53). The rAAV2 was isolated from the 40%-60% interface with a syringe.
Quantification of packaged DNA.
Recovery of virus in terms of genome copies was determined by DNA hybridization. Ten microliters of each sample was incubated in 100 μl DNase I digestion buffer (10 mM Tris, pH 7.5, 10 mM MgCl2, and 50 units/ml of DNase I) and incubated at 37°C for 1 h. The DNase digestion was stopped by the addition of 4 μl of 0.5 M EDTA. One hundred twenty microliters of proteinase K solution (1 M NaCl, 100 μg/ml proteinase K, 1% Sarkosyl) was then added and incubated at 50°C for 2 h; 250 μl of phenol-chloroform was added to the digestion mixture, mixed, and centrifuged at 16,000 × g for 5 min; and 1 μl of 20-mg/ml glycogen, 62.5 μl of 10 M ammonium acetate, and 2.5 volumes of 100% ethanol were added to the aqueous phase to precipitate viral DNA. Samples were placed on dry ice for 1 h and then centrifuged at 16,000 × g for 20 min to pellet the viral DNA. The DNA pellet was resuspended in 200 μl of Tris-EDTA. After 0.5 M NaOH digestion for 10 min, aliquots of the samples were applied to a dot blot manifold. The denatured viral DNA was then blotted onto a nylon membrane and probed with a P32-labeled probe against the luciferase transgene.
Infectious-center assay.
Infectivity assays were carried out on the C12 cell line (11, 12), a HeLa cell line with the wt AAV2 coding region integrated with a neomycin resistance gene for selection. Cells were seeded at a density of 5 × 104/well of a 48-well plate. The cells were infected the following day with various dilutions of the vector of interest, along with an adenovirus multiplicity of infection of 20. The cells were harvested 42 h postinfection and washed with 0.5 ml of 1× phosphate-buffered saline (PBS), and the wash was added to a collection tube. One hundred twenty microliters of Trypsin plus EDTA was then added to each well, the cells were incubated at 37°C for 6 to 8 min, 0.5 ml of medium was added to each well, and the cells were removed. The cells were then applied to a Hybond Nylon membrane via an ICA manifold using a vacuum. The membrane was allowed to dry and was then applied to Whatman paper saturated in denaturation solution (1.5 M NaCl and 0.5 M NaOH) for 8 min. The membrane was applied to Whatman paper saturated in neutralization solution (0.5 M NaCl and 0.5 M Tris-HCl, pH 7.5) for 8 min. The membranes were then UV cross-linked and probed overnight with a transgene-specific probe. Based on the number of dots (cells with active replication of vector genome) and the number of total viral genomes used to infect the cells, an infectious-to-noninfectious-particle ratio could be determined.
Native dot blot Western assay to determine accessibility of N termini.
A native dot blot Western assay was carried out as previously described (6). Briefly, 5 × 109 iodixanol-purified capsids were exposed to a temperature of 60°C for 5 min. The heat-treated samples were then incubated on ice for 5 min and transferred to a Hybond-ECL membrane (Amersham) by using a vacuum dot blotter. The membranes were blocked for 1 h in 1× PBS containing 10% milk powder (blocking solution) and then incubated for 1 h with the monoclonal antibody (MAb) A20, A1, or B1 diluted 1:10 in blocking solution. The membranes were washed six times with 1× PBS for 10 min each time, followed by incubation with a peroxidase-coupled goat anti-mouse antibody for 1 h. After six washes with PBS for 10 min each time, the proteins were visualized by using a chemiluminescence detection kit (Pierce).
Transduction assays.
HeLa cells were plated at a density of 4 × 104/well of a 96-well plate the day of infection. The HeLa cells were then infected at 1 × 104 viral genomes (vg)/cell with each vector produced in this study. Luciferase activity was measured 24 h postinfection in accordance with the manufacturer's instructions (Promega). Luciferase activity was measured with a Tropix TR717 automated plate reader.
Confocal-microscopy imaging of AAV virions.
HeLa cells (5 × 104/well) were plated on polylysine-coated 12-mm glass coverslips 24 h before infection. Recombinant virions of AAV2, AAV2-ala, AAV2-cys, or AAV2-trp were added to the cell media (3 × 104 vg/cell) and incubated in the presence of 40 mM ALLN (Calbiochem). No virus was added to control wells. After 4 h, the cells were washed three times with PBS and then fixed with 2% paraformaldehyde for 15 min at room temperature. The cells were then permeabilized with 0.1% Triton X-100 in PBS for 5 min at room temperature. Following four washes with PBS, the permeabilized cells were blocked with immunofluorescence wash buffer (IFWB) (20 mM Tris, pH 7.5, 137 mM NaCl, 3 mM KCl, 1.5 mM MgCl2, 5 mg/ml bovine serum albumin, 0.05% Tween) for 30 min at room temperature. The cells were incubated with primary antibody to detect intact capsids (MAb A20), diluted 1:10 in IFWB, for 1 h at 37°C. The cells were then incubated in secondary antibody, diluted 1:1,250 in IFWB (anti-mouse Alexa Fluor 488; Molecular Probes), for 1 h at 37°C. After six washes in PBS, coverslips were mounted cell side down on glass slides with mounting medium (Prolong antifade Gold with DAPI [4′,6′-diamidino-2-phenylindole]; Molecular Probes). Images were captured on a Leica SP2 AOBS upright laser scanning confocal microscope and processed using Adobe Photoshop.
EM.
Heparin column-purified virus from iodixanol gradients was placed on a 400-mesh glow-discharged carbon grid by inversion on a 20-μl drop of virus. The grid was washed three times in 20 liters of PBS for 1 min. The virus was then stained for 1 min with 2% uranyl acetate and visualized by using a Zeiss EM 910 electron microscope.
Phospholipase assay.
Viral particles (1 × 1010) were used in a phospholipase assay to assess PLA2 activity on the surfaces of rAAV2 fivefold pore mutants and Vp3-only virions. Assays were performed utilizing the sPLA2 Assay Kit from Caymen Chemical. The assay was carried out in accordance with the manufacturer's protocol but with extended incubation periods between absorbance (405 nm) readings.
RESULTS
Vp1 fusion constructs.
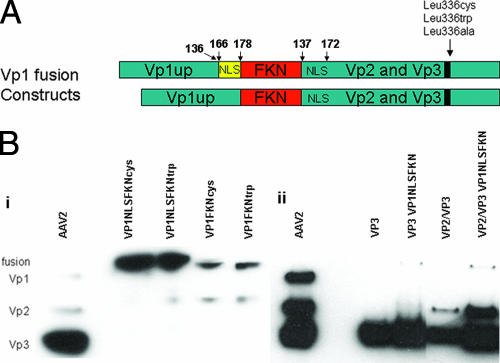
As previously described (6), mutagenesis of Leu336 led to a dramatic decrease in infectivity. This decrease has been attributed to inhibition of Vp1 N-terminus exposure. To further explore what role the mutations play in AAV infection, we took a unique approach to determine if it is possible to rescue/enhance the infectivity of these mutant virions by assembling rAAV virions with surface-exposed Vp1 (Vp1 fusion proteins). A study conducted by Warrington et al. established that it is possible to fuse FKN (8 kDa), Lep (18 kDa), and GFP (28 kDa) to the N terminus of Vp2 and have them surface displayed on the virion (43). We utilized the Vp2AFKN construct kindly provided to us by K. H. Warrington and fused the unique N terminus of Vp1 with and without an NLS to the N terminus of FKN. The NLS (166PARKRLN172) used in the Vp1 fusion protein in this study was previously determined to be located, and functional, in both Vp1 and Vp2 capsid proteins (19). Briefly, the ATG start codon on FKN was mutated to CTG in order to initiate translation from the Vp1 start site only, producing the Vp1FKN or Vp1NLSFKN protein (see Materials and Methods for details). The Vp1 fusion constructs used in this study are shown in Fig. 2A, and a Western blot verifying that each Vp1 fusion construct was produced from the corresponding plasmid at the correct size is shown in Fig. 2B.
FIG. 2.
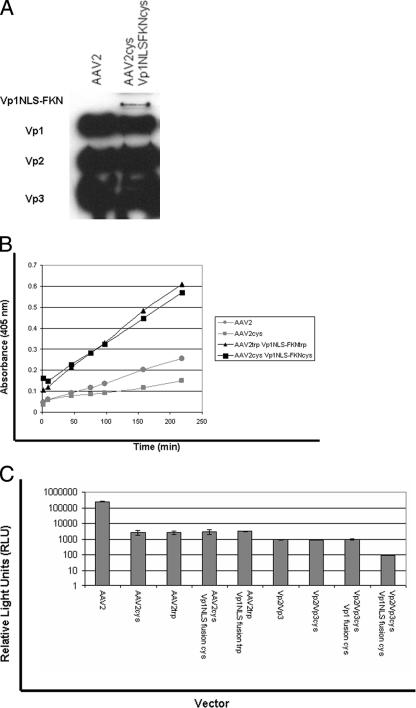
(A) Schematic representation of the Vp1FKN and Vp1NLSFKN fusion proteins generated for this study. The numbers above the constructs represent amino acid sequence. (B) Western blots of (i) cell lysate that had been previously transfected with the Vp1 fusion proteins and (ii) purified AAV2 virions with and without the Vp1NLSFKN fusion protein. The MAb B1 was used as the primary antibody in the Western blot analysis of the capsid proteins.
In a previous study, fusion proteins of various sizes were engineered to the N terminus of Vp2 and found to be located on the surfaces of AAV2 virions (43). It was determined that when transfected at higher concentrations, the largest fusion protein (Vp2AGFP) negatively impacted physical and infectious-particle titers. Thus, the Vp1 fusion proteins, being similar in size to Vp2AGFP, were titrated at a 1:20 ratio, as specified by Warrington et al. (43), to the helper plasmids (helper plasmids were derived from pXR2 expressing Vp2/Vp3 and Vp3 only) used when transfecting 293 cells for rAAV2 production. Western blot data (Fig. 2B) illustrate that the Vp1 fusion proteins migrate above the wt Vp1 protein as predicted and can be incorporated into wt virions, along with virions composed of Vp3-only, Vp2/Vp3-only, and Leu336 mutant virions. It is apparent that the Vp1 fusion proteins are incorporated into the virion, using this titration ratio, at low levels as predicted and consistent with the results of Warrington et al. (43).
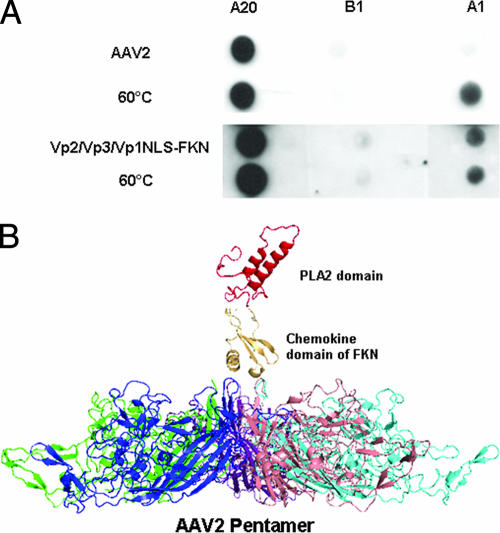
To determine the abilities of the Vp1 fusion constructs to function in the context of a virion composed of wt capsid subunits, they were incorporated into Vp2/Vp3 and Vp3-only particles. We chose these particles for the characterization studies because they are noninfectious when they lack the Vp1 subunit. The Vp2/Vp3/Vp1NLSFKN virion was analyzed via a native dot blot assay to determine if the Vp1NLSFKN protein was surface exposed. Viral-genome-containing particles (109) were adsorbed to a nylon membrane and probed with MAbs A20, B1, and A1, which bind to intact virions, unassembled virions, and Vp1 N termini, respectively. As a control, wt AAV2 virions were used (positive A20 signal) and heated to 60°C prior to adsorption to the membrane to show Vp1 exposure (positive A1 signal). As illustrated in Fig. 3, it is clearly evident that that the Vp1NLSFKN domain is surface exposed when incorporated into the Vp2/Vp3 virion. When the Vp2/Vp3/Vp1NLSFKN virion was heated to 60°C, an increase in the A1 antibody signal was not detected, which was unlike the wt AAV2 control and further supports surface exposure of this chimeric protein when incorporated into viral particles. Together, these data suggest that all of the Vp1NLSFKN N termini are surface exposed on the virion. Due to low levels of Vp1NLSFKN protein incorporated into the virion, a longer exposure time was needed to detect the A1 signal, while the B1 signal that detects unassembled capsids remained just above background level. A schematic of the Vp1 fusion protein on the surface of the AAV2 virion is shown Fig. 3B.
FIG. 3.
(A) Native dot blot Western analysis of purified AAV2 and Vp2/Vp3/Vp1NLSFKN. A20 (MAb to intact AAV2 virions), B1 (MAb to individual capsid proteins), and A1 (MAb to the unique N terminus of VP1) were used to assess the surface exposure of wt VP1 for AAV2 when heated to 60°C and Vp1NLSFKN when incorporated into Vp2/Vp3-only particles at room temperature. (B) Schematic representation of the surface-exposed domains of the Vp1 fusion proteins. This is not a predicted structure but was compiled to provide a visualization of the possible juxtaposition of the FKN and PLA2 protein regions above the AAV2 fivefold pore region. It was generated from the crystal structure of the AAV2 Vp3 protein (Protein Data Bank [PDB] identifier 1LP3), the structure of the chemokine domain of FKN (PDB identifier 1F2L), and a homologous model of the AAV2 PLA2 domain generated by sequence comparison to the PLA2 domain structure of bee venom using the SWISS MODEL algorithm (33a; http://www.expasy.org). The FKN and PLA2 domain structures were visually “docked” above the fivefold pore in the AAV2 Vp3 pentamer in the PyMol program (W. L. DeLano, The PyMol molecular graphics system, DeLano Scientific, San Carlos, CA, 2002).
Phospholipase assay to measure PLA2 activities of the Vp1 fusion proteins.
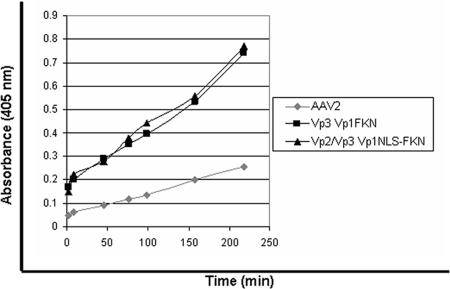
To determine if the PLA2 domain in the Vp1 fusion proteins is functional in the context of the virus, a phospholipase assay was performed on the Vp3/Vp1FKN and Vp2/Vp3/Vp1NLSFKN virions. Wt AAV2 virions were used as a control in this experiment. As depicted in Fig. 4, Vp1 fusion protein-containing virions showed PLA2 activity, while the wt AAV2 virion did not, due to the fact that the Vp1 N termini in wt AAV2 virions are located within the virion. As previously described, wt AAV displays lipase activity in vitro, but only after being heated further, supporting the idea that the Vp1 fusion proteins are surface exposed on the virion during assembly.
FIG. 4.
PLA2 assay. The PLA2 assay was carried out at room temperature, using AAV2 as the negative control, to assess the surface exposure and functionality of the Vp1 fusion proteins according to the manufacturer's protocol.
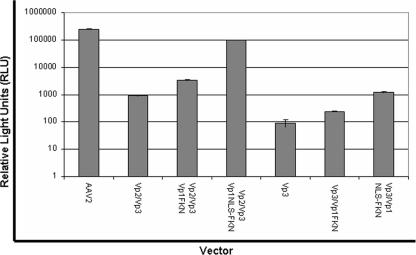
The Vp1 fusion proteins aid in infectivity.
Two sets of virus particles (set 1, Vp3 only, Vp3/Vp1FKN, and Vp3/Vp1NLSFKN, and set 2, Vp2/Vp3 only, Vp2/Vp3/Vp1FKN, and Vp2/Vp3/Vp1NLSFKN) encapsidating the firefly luciferase transgene cassette were produced. HeLa cells were infected with the aforementioned viral particles at 104 vg/cell, and their ability to transduce cells was assessed via luciferase expression in relative light units (Fig. 5). The Vp1 fusion proteins were observed to confer infectivity on the noninfectious Vp3- and Vp2/Vp3-only particles. There was a three- to fivefold increase in transduction when the Vp1FKN protein was incorporated into Vp3- or Vp2/Vp3-only expression cassettes. We observed a 10- to 100-fold increase in transduction when Vp1NLSFKN was incorporated into the Vp3-only and the Vp2/Vp3-only virions, respectively. While these data strongly suggested that the PLA2 activity can rescue AAV virion infectivity, it also clearly established that PLA2 activity alone is not enough to confer optimum infectivity. The NLS must be surface exposed, along with the Vp1 N terminus, for a successful infection to occur.
FIG. 5.
Luciferase transduction assay for comparison of wt AAV2 virions and virions containing VP1 fusion proteins in substitution for wt VP1. HeLa cells were infected at 1 × 104 vg/cell with each vector (n = 4). Luciferase activity was then measured 24 h postinfection in accordance with the manufacturer's instructions (Promega).
Initial characterization of the fivefold pore mutants.
Two parvovirus studies reported that mutagenesis of the residues making up the fivefold pore led to a 2 to 4 log10 reduction in infectivity (43). The data generated by Bleker et al. demonstrated that a majority of the fivefold pore mutants, including the conserved Leu336, inhibited Vp1 N-terminus exposure, which explained the decrease in infectivity. Therefore, we targeted the conserved Leu336 residue for mutagenesis (Leu336 to Ala, Cys, and Trp) to determine if the addition of Vp1NLSFKN can enhance/restore infectivity to wt AAV2 levels. Utilizing the dot blot method to titer each mutant, we established that AAV2-ala, AAV2-cys, and AAV2-trp consistently produced wt, 2- to 3-fold-lower, and 7- to 10-fold-lower levels of genome-containing particles, respectively, than vectors produced from wt AAV2 capsid proteins (data not shown). Similar to the data reported for the MVM Leu172Trp mutation, the AAV2-trp mutant seemed to sustain a packaging defect, but not to the same level as that reported for the autonomous parvovirus.
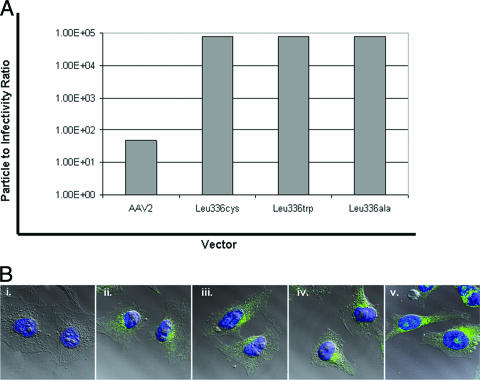
Infectious-center assays were performed on each of the mutants to determine the infectious-to-noninfectious-particle ratio for each vector, as described in Materials and Methods. These data show a 3 log10 difference in infectivity between AAV2 and the Leu336 mutants (Fig. 6A). The infectious-to-noninfectious-particle ratios were 1:48 for wt AAV2 and ∼1:75,000 for the Leu336 mutants. Western blot analysis determined that the purified Leu336 mutant virions were capable of assembly, as well as incorporating all of the capsid subunits into the virion at wt ratios (data not shown) (6, 17). To further characterize at what step the Leu336 mutants were defective in infectivity, confocal-microsopy experiments were carried out to determine if the Leu336 mutant virions were capable of binding and internalizing into cells compared to wt AAV2 virions (Fig. 6B). The A20 antibody was used to determine the location of the virions 4 hours after HeLa cells were infected with 3 × 104 vg/cell. Using this approach, the Leu336 mutant virions were observed to be capable of binding, internalizing into, and trafficking to the perinuclear region within the HeLa cells as efficiently as wt AAV2 virions. Taken together, the above-mentioned data imply that the reduced infectivity of the Leu336 mutants is not due to the lack of minor capsid components, such as Vp1, or the ability of the virions to bind and internalize into cells.
FIG. 6.
(A) Infectious-center assay data to assess the particle-to-infectivity ratio between wt AAV2 virions and the Leu336 mutant virions. (B) Confocal-microscopy images of (i) A20 negative control and (ii) AAV2-ala-, (iii) AAV2-cys-, (iv) AAV2-trp-, and (v) wt AAV2-infected HeLa cells. MAB A20 was used to assess the locations of the virions 4 h postinfection.
Leu336 mutants have dominant mutations.
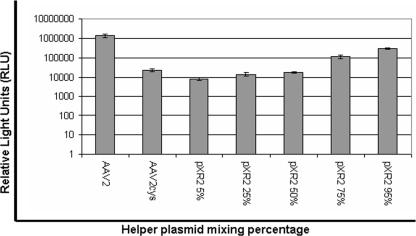
To determine if the fivefold pore mutants have a dominant mutation, we mixed the AAV2-cys and wt helper plasmids at different ratios to one another (1:19, 1:3, 1:1, 3:1, and 19:1). Luciferase reporter assays were then carried out to determine the effects the various concentrations of the AAV2-cys capsid subunits incorporated into the virion have on the overall transduction profile. HeLa cells were infected with 104 vg/cell in triplicate and assayed for luciferase activity 24 h postinfection. The trend was as expected; as the amount of wt helper (pXR2) was increased, transduction levels also increased (Fig. 7). Interestingly, 75%:25% and 95%:5% ratios of pXR2 to AAV2-cys helpers were needed to increase transduction above mutant virus levels and within a log unit of AAV2. This lends further support to the idea that the Leu336 mutants have dominant mutations when incorporated into capsid.
FIG. 7.
Luciferase transduction assay for comparison of wt AAV2 virions and chimeric virions composed of various concentrations (95%, 75%, 50%, 25%, and 5%) of the Leu336-cys capsid protein (n = 4).
Transmission EM analysis of the mutant virions.
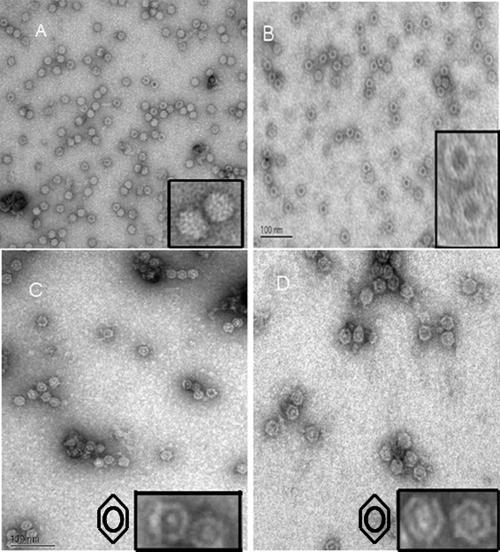
Negative-stain transmission EM analysis was done on each of the Leu336 mutant virions to visualize their integrity. A field from each negative-stain micrograph is displayed in Fig. 8 with two virions from each respective field enlarged to enable closer inspection. All the mutants assembled intact capsids. The AAV2-cys and AAV2-trp mutants were observed to have altered negative-stain permeability compared to wt AAV2 virions. While the staining pattern for the AAV2-ala mutant was similar to that expected for empty capsids (a darkly stained center), the AAV2-cys and AAV2-trp mutant capsids showed a distinct staining pattern with a dark ring around the center of the capsid rather than at the center. However, data generated from dot blot/Southern blot analysis, in addition to the fact that each virus was removed from a region of the iodixanol step gradient, where genome-containing particles have been determined to reside, strongly suggests that the virions depicted in the EM should contain genomes and not be represented by empty particles observed across the entire micrograph. Negative-stain transmission EM analysis of empty AAV2-ala mutant particles showed a staining pattern identical to those of the genome-containing particles depicted in Fig. 8 (data not shown). Although confocal-microscopy analysis supported normal trafficking of the fivefold pore mutants, EM data suggest that the structure/conformation of the fivefold pore mutant virions differs from that of wt AAV2 virions.
FIG. 8.
Negative-stain EM images of (A) wt AAV2, (B) AAV2-ala, (C) AAV2-cys, and (D) AAV2-trp that identified the unique staining pattern of the Leu336 mutant virions. The insets are magnified close-ups of the AAV2 virions. Symbols in panels C and D represent the staining pattern/profile of the fivefold mutant virlons.
Vp1NLSFKN incorporation into Leu336 mutant virions.
To determine if the Vp1NLSFKN protein could assist in the infectious entry pathway of the Leu336 mutant virions, we first needed to confirm that it was incorporated into the mutant virions. The Western blot depicted in Fig. 9A illustrates that the Vp1NL-FKN protein is incorporated into Leu336 mutant virions. Exposure time for the Western blot was extended to detect the signal from the Vp1NLSFKN protein, as also described for wt incorporation (Fig. 2B). In addition to the capsid composition, we next had to determine if the PLA2 domain is functional and surface exposed. A PLA2 assay was carried out on AAV2, AAV2-cys, AAV2-cys Vp1NLSFKNcys, and AAV2-trp Vp1NLSFKNtrp to determine if Vp1NLSFKN is functional and surface exposed. The PLA2 assay depicted in Fig. 9B shows that AAV2 and AAV2-cys do not have PLA2 activity, while the AAV2-cys Vp1NLSFKNcys and AAV2-trp Vp1NLSFKNtrp virions display PLA2 activity without exposure to heat. Therefore, the Vp1NLSFKN protein is surface exposed and functional when incorporated into the Leu336 mutant virions. However, in contrast to the data presented in Fig. 5, the Vp1NLSFKN protein incorporated into the Leu336 mutant virions did not enhance/restore infectivity compared to their respective parental mutant controls (Fig. 9C), suggesting an additional role of the fivefold pore.
FIG. 9.
(A) Western blot of purified Leu336 mutant containing the Vp1NLSFKN fusion protein that was previously transfected 1:20 to the helper plasmid. (B) PLA2 assay. The PLA2 assay was carried out at room temperature, using AAV2 and AAV2-cys as negative controls to assess the surface exposure and functionality of the Vp1 fusion proteins when incorporated into the Leu336 mutant virions according to the manufacturer's protocol. (C) Luciferase transduction assay for comparison of wt AAV2 and Leu336 mutant virions containing the VP1 fusion proteins and Leu336 mutant virions containing VP1 fusion proteins in substitution for wt VP1 (n = 4).
DISCUSSION
Two of the most prominent features of the surface topology of the AAV capsid are the threefold proximal peaks and the cylindrical pore at the fivefold axis of symmetry (Fig. 1A). The fivefold pore connects the outer surface of the capsid with the inner surface and is large enough to allow transit of nucleic acids and possibly linear protein domains, such as the Vp1 and Vp2 N termini. It is thought that after transit through the pore, the Vp1 N terminus must fold into its functional conformation (24). Based on parvovirus capsid amino acid alignments and data generated from previous parvovirus fivefold pore studies (24), we decided to focus our study on the conserved leucine residue 336 at the base of the pore. Bleker et al. determined that mutagenesis of single amino acids lining the pore inhibited Vp1 surface exposure. The purpose of this study was to further explore the role of the fivefold pore in the life cycle of AAV2. We decided to take a unique approach to determine the significance of the pore by generating Vp1 fusion constructs that would place the unique N terminus of Vp1 on the outside of the virus, similar to the autonomous parvovirus B19, and to determine if the Vp1 fusion proteins are functional in the context of the wt and Leu336 mutant virions.
Based on a number of studies, it has been proposed that AAV requires endosomal acidification to escape from the late endosome and traffic to the nucleus (16, 20, 50, 51). These studies also suggest that prior to escaping the endosome, the virion must undergo conformational changes, leading to the exposure of the unique N terminus of Vp1 and the N terminus of Vp2 required for endososmal escape and nuclear entry (6, 9, 19, 33, 35). The unique N terminus of Vp1 contains a conserved PLA2 domain (8, 15, 18, 28, 52) that is essential for infectivity and is thought to be required for endosomal escape. A suggestion that the N termini of Vp1 are located within the virion and become surface exposed naturally within the cell, via an induced conformational change, is consistent with the observation that while intact capsids do not have PLA2 activity, heat or acidic-pH treatment of virions elicits this function (37, 52). Basic amino acid clusters, which are thought to control the nuclear import of incoming virions, are also contained within the unique Vp1 N terminus (27, 41) and the N-terminal amino acid stretch (166PARKRLN172) contained within an overlapping region of Vp1 and Vp2 (19). It has been hypothesized that the fivefold pore is used as the portal for Vp1 N-terminus translocation to the surface of the virion, based on mutagenesis studies (6). This previous study demonstrated that mutagenesis of single amino acids making up the pore led to the inhibition of Vp1 N-terminus exposure and a substantial decrease in infectivity (Fig. 6A). Taken together, the Vp1 fusion proteins were designed, with all of the above in mind, with the intention to surface expose the PLA2 domain with and without the NLS. Insertions of foreign epitopes at the N termini of Vp1 and Vp2 have been reported to present these proteins on the surfaces of mutant virions, as indicated by their accessibility to antibodies (43, 46). This suggests that these large protein insertions become exposed during the assembly process because it is unlikely, considering their size, that they would internalize into the virion. The Vp1 fusion proteins reconfirmed the essential components needed to carry out a successful infection, along with determining if infectivity of the fivefold pore mutants, which have demonstrated inhibition of Vp1 N-terminus exposure, can be enhanced/restored.
Vp1 fusion proteins are functional in the context of AAV2 virions.
The Vp1 fusion proteins were successfully incorporated and functional in wt capsids composed of Vp3 only and Vp2/Vp3 based on Western blot analysis and PLA2 data (Fig. 2B and 4). Infectivity was enhanced or restored for these wt capsids, which are normally noninfectious because of their deficiency of wt Vp1 proteins. In the case of the Vp3 capsids, 3- and 10-fold increases in transduction were observed when Vp1FKN and Vp1NLSFKN were incorporated. Incorporation of Vp1FKN and Vp1NLSFKN into the Vp2/Vp3 virions led to a 5- and a 100-fold increase, respectively. The above data are in agreement with a previous Vp1/Vp2 NLS complementation study (19), which demonstrated that the PLA2 activity on Vp1 is not enough to confer infectivity. The NLS must be surface exposed, along with the Vp1 N terminus (Vp1NLSFKN), for a successful infection to occur. In addition, these data also demonstrate that capsids containing Vp2 transduce cells more efficiently than in its absence (Fig. 5, Vp3/Vp1NLSFKN and Vp2/Vp3/Vp1NLSFKN), which agrees with data generated in the Vp1/Vp2 NLS complementation study that identified NLS functional redundancy between Vp1 and Vp2 (19). However, when Vp1NLSFKN was incorporated into the Leu336 mutant capsids, infectivity was not enhanced/restored, even though the Vp1NLSFKN was surface exposed and displayed PLA2 activity (Fig. 9B). These data strongly suggest that inhibition of Vp1 N termini by the fivefold pore mutants is not the only factor involved in decreased infectivity. To further support this statement, negative-stain transmission EM analysis of the Leu336 mutant capsids showed that they appeared to have altered permeability compared to wt AAV2 capsids (Fig. 8), suggesting that they are structurally/conformationally distinct from wt AAV2 capsids. In addition, virions generated from mixing low concentrations (5% and 25%) of Leu336 mutant helper plasmids with wt helper plasmid during transfection led to a 5- to 10-fold decrease in infectivity. This suggests that the Leu336 mutants have dominant mutations and that incorporation at low levels into wt capsids can potentially result in virion conformation/structure changes to impact infectivity. Together, these observations suggest that the Leu336 mutants likely cause global structural changes to the virion, directly impacting downstream conformational changes essential for infectivity and do not only have local effects within the pore, as previously reported (6).
This study establishes that it is possible to generate AAV virions with surface-displayed functional proteins or domains that can augment infectivity and provides further supporting data for the significance of surface exposing Vp1 N termini in conjunction with the NLS during infection. It is currently unknown why the Leu336 mutants are defective for infectivity. However, data generated in this study have led to the proposal that the fivefold mutations cause structural alterations to the virion and/or prevent downstream conformational changes essential for proper egress of the virion after it enters the cell. If the fivefold pore is the location for Vp1 N-terminus exposure, the mechanism by which this occurs has yet to be elucidated. Understanding this mechanism is of high priority. Once a thorough understanding of the infectious entry pathway is established, it may be possible to fuse and utilize other “lipid-penetrating” proteins/domains or other proteins advantageous to Vp1 and or Vp2 for the generation of better AAV vectors for gene therapy.
Acknowledgments
We thank K. H. Warrington for providing the Vp2AFKN plasmid for use in this study.
This study was supported by PO1 HL59412 HL51811, GM059299, HL066973, and P30 DK34987 and P30 Dk065988.
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.Agbandje, M., S. Kajigaya, R. McKenna, N. S. Young, and M. G. Rossmann. 1994. The structure of human parvovirus B19 at 8 Å resolution. Virology 203:106-115. [DOI] [PubMed] [Google Scholar]
- 2.Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and C. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155-171. [DOI] [PubMed] [Google Scholar]
- 3.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 4.Asokan, A., J. B. Hamra, L. Govindasamy, M. Agbandje-McKenna, and R. J. Samulski. 2006. Adeno-associated virus type 2 contains an integrin α5β1 binding domain essential for viral cell entry. J. Virol. 80:8961-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atchison, R. W., B. C. Casto, and W. M. Hammon. 1965. Adenovirus-associated defective virus particles. Science 149:754-756. [DOI] [PubMed] [Google Scholar]
- 6.Bleker, S., F. Sonntag, and J. A. Kleinschmidt. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 79:2528-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buller, R. M., J. E. Janik, E. D. Sebring, and J. A. Rose. 1981. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol. 40:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canaan, S., Z. Zadori, F. Ghomashchi, J. Bollinger, M. Sadilek, M. E. Moreau, P. Tijssen, and M. H. Gelb. 2004. Interfacial enzymology of parvovirus phospholipases A2. J. Biol. Chem. 279:14502-14508. [DOI] [PubMed] [Google Scholar]
- 9.Carreira, A., M. Menendez, J. Reguera, J. M. Almendral, and M. G. Mateu. 2004. In vitro disassembly of a parvovirus capsid and effect on capsid stability of heterologous peptide insertions in surface loops. J. Biol. Chem. 279:6517-6525. [DOI] [PubMed] [Google Scholar]
- 10.Chejanovsky, N., and B. J. Carter. 1989. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology 173:120-128. [DOI] [PubMed] [Google Scholar]
- 11.Clark, K. R., F. Voulgaropoulou, D. M. Fraley, and P. R. Johnson. 1995. Cell lines for the production of recombinant adeno-associated virus. Hum. Gene Ther. 6:1329-1341. [DOI] [PubMed] [Google Scholar]
- 12.Clark, K. R., F. Voulgaropoulou, and P. R. Johnson. 1996. A stable cell line carrying adenovirus-inducible rep and cap genes allows for infectivity titration of adeno-associated virus vectors. Gene Ther. 3:1124-1132. [PubMed] [Google Scholar]
- 13.Cotmore, S. F., M. A. D'Abramo, Jr., C. M. Ticknor, and P. Tattersall. 1999. Controlled conformational transitions in the MVM virion expose the VP1 N-terminus and viral genome without particle disassembly. Virology 254:169-181. [DOI] [PubMed] [Google Scholar]
- 14.Di Pasquale, G., B. L. Davidson, C. S. Stein, I. Martins, D. Scudiero, A. Monks, and J. A. Chiorini. 2003. Identification of PDGFR as a receptor for AAV-5 transduction. Nat. Med. 9:1306-1312. [DOI] [PubMed] [Google Scholar]
- 15.Dorsch, S., G. Liebisch, B. Kaufmann, P. von Landenberg, J. H. Hoffmann, W. Drobnik, and S. Modrow. 2002. The VP1 unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J. Virol. 76:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douar, A. M., K. Poulard, D. Stockholm, and O. Danos. 2001. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J. Virol. 75:1824-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farr, G. A., and P. Tattersall. 2004. A conserved leucine that constricts the pore through the capsid fivefold cylinder plays a central role in parvoviral infection. Virology 323:243-256. [DOI] [PubMed] [Google Scholar]
- 18.Farr, G. A., L. G. Zhang, and P. Tattersall. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. USA 102:17148-17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grieger, J. C., S. Snowdy, and R. J. Samulski. 2006. Separate basic region motifs within the adeno-associated virus capsid proteins are essential for infectivity and assembly. J. Virol. 80:5199-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen, J., K. Qing, and A. Srivastava. 2001. Adeno-associated virus type 2-mediated gene transfer: altered endocytic processing enhances transduction efficiency in murine fibroblasts. J. Virol. 75:4080-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 22.Kashiwakura, Y., K. Tamayose, K. Iwabuchi, Y. Hirai, T. Shimada, K. Matsumoto, T. Nakamura, M. Watanabe, K. Oshimi, and H. Daida. 2005. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J. Virol. 79:609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King, J. A., R. Dubielzig, D. Grimm, and J. A. Kleinschmidt. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kronenberg, S., B. Bottcher, C. W. von der Lieth, S. Bleker, and J. A. Kleinschmidt. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 79:5296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kronenberg, S., J. A. Kleinschmidt, and B. Bottcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llamas-Saiz, A. L., M. Agbandje-McKenna, W. R. Wikoff, J. Bratton, P. Tattersall, and M. G. Rossmann. 1997. Structure determination of minute virus of mice. Acta Crystallogr, D 53:93-102. [DOI] [PubMed] [Google Scholar]
- 27.Lombardo, E., J. C. Ramirez, J. Garcia, and J. M. Almendral. 2002. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J. Virol. 76:7049-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lupescu, A., C. T. Bock, P. A. Lang, S. Aberle, H. Kaiser, R. Kandolf, and F. Lang. 2006. Phospholipase A2 activity-dependent stimulation of Ca2+ entry by human parvovirus B19 capsid protein VP1. J. Virol. 80:11370-11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lusby, E., K. H. Fife, and K. I. Berns. 1980. Nucleotide sequence of the inverted terminal repetition in adeno-associated virus DNA. J. Virol. 34:402-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lux, K., N. Goerlitz, S. Schlemminger, L. Perabo, D. Goldnau, J. Endell, K. Leike, D. M. Kofler, S. Finke, M. Hallek, and H. Buning. 2005. Green fluorescent protein-tagged adeno-associated virus particles allow the study of cytosolic and nuclear trafficking. J. Virol. 79:11776-11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenna, R., N. H. Olson, P. R. Chipman, T. S. Baker, T. F. Booth, J. Christensen, B. Aasted, J. M. Fox, M. E. Bloom, J. B. Wolfinbarger, and M. Agbandje-McKenna. 1999. Three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity. J. Virol. 73:6882-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qing, K., C. Mah, J. Hansen, S. Zhou, V. Dwarki, and A. Srivastava. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 5:71-77. [DOI] [PubMed] [Google Scholar]
- 33.Reguera, J., A. Carreira, L. Riolobos, J. M. Almendral, and M. G. Mateu. 2004. Role of interfacial amino acid residues in assembly, stability, and conformation of a spherical virus capsid. Proc. Natl. Acad. Sci. USA 101:2724-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Schwede T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpson, A. A., P. R. Chipman, T. S. Baker, P. Tijssen, and M. G. Rossmann. 1998. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure 6:1355-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonntag, F., S. Bleker, B. Leuchs, R. Fischer, and J. A. Kleinschmidt. 2006. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 80:11040-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suikkanen, S., M. Antila, A. Jaatinen, M. Vihinen-Ranta, and M. Vuento. 2003. Release of canine parvovirus from endocytic vesicles. Virology 316:267-280. [DOI] [PubMed] [Google Scholar]
- 38.Summerford, C., J. S. Bartlett, and R. J. Samulski. 1999. αVβ5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med. 5:78-82. [DOI] [PubMed] [Google Scholar]
- 39.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsao, J., M. S. Chapman, M. Agbandje, W. Keller, K. Smith, H. Wu, M. Luo, T. J. Smith, M. G. Rossmann, R. W. Compans, et al. 1991. The three-dimensional structure of canine parvovirus and its functional implications. Science 251:1456-1464. [DOI] [PubMed] [Google Scholar]
- 41.Vihinen-Ranta, M., D. Wang, W. S. Weichert, and C. R. Parrish. 2002. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 76:1884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walters, R. W., M. Agbandje-McKenna, V. D. Bowman, T. O. Moninger, N. H. Olson, M. Seiler, J. A. Chiorini, T. S. Baker, and J. Zabner. 2004. Structure of adeno-associated virus serotype 5. J. Virol. 78:3361-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warrington, K. H., Jr., O. S. Gorbatyuk, J. K. Harrison, S. R. Opie, S. Zolotukhin, and N. Muzyczka. 2004. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J. Virol. 78:6595-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weichert, W. S., J. S. Parker, A. T. Wahid, S. F. Chang, E. Meier, and C. R. Parrish. 1998. Assaying for structural variation in the parvovirus capsid and its role in infection. Virology 250:106-117. [DOI] [PubMed] [Google Scholar]
- 45.Wu, H., and M. G. Rossmann. 1993. The canine parvovirus empty capsid structure. J. Mol. Biol. 233:231-244. [DOI] [PubMed] [Google Scholar]
- 46.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, W., K. H. Warrington, Jr., P. Hearing, J. Hughes, and N. Muzyczka. 2002. Adenovirus-facilitated nuclear translocation of adeno-associated virus type 2. J. Virol. 76:11505-11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie, Q., and M. S. Chapman. 1996. Canine parvovirus capsid structure, analyzed at 2.9 Å resolution. J. Mol. Biol. 264:497-520. [DOI] [PubMed] [Google Scholar]
- 50.Yan, Z., R. Zak, G. W. Luxton, T. C. Ritchie, U. Bantel-Schaal, and J. F. Engelhardt. 2002. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 76:2043-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan, Z., R. Zak, Y. Zhang, W. Ding, S. Godwin, K. Munson, R. Peluso, and J. F. Engelhardt. 2004. Distinct classes of proteasome-modulating agents cooperatively augment recombinant adeno-associated virus type 2- and type 5-mediated transduction from the apical surfaces of human airway epithelia. J. Virol. 78:2863-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291-302. [DOI] [PubMed] [Google Scholar]
- 53.Zolotukhin, S., B. J. Byrne, E. Mason, I. Zolotukhin, M. Potter, K. Chesnut, C. Summerford, R. J. Samulski, and N. Muzyczka. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973-985. [DOI] [PubMed] [Google Scholar]