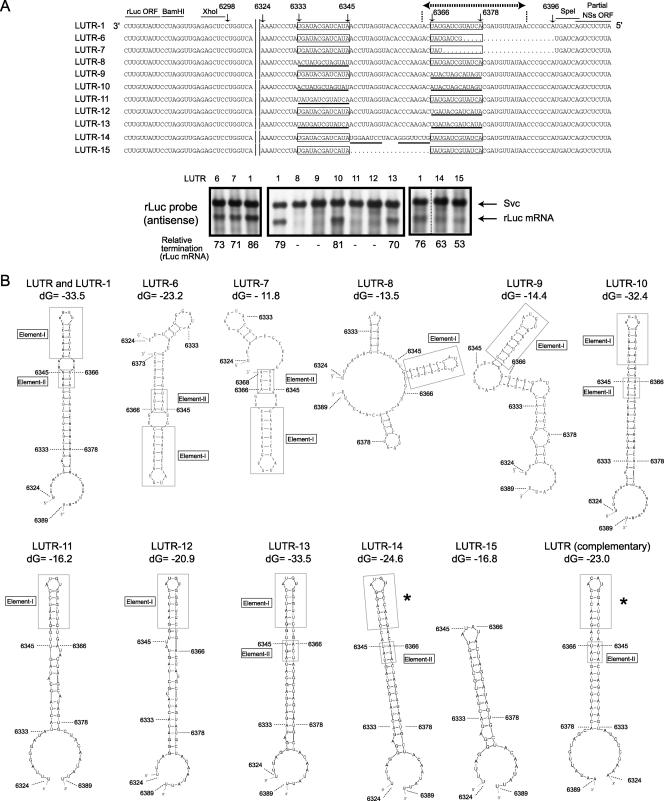
FIG. 10.
Analysis of L mRNA transcription termination control. (A) L-5′ UTR sequences in LUTR-1 and other mutants are shown. The numbers represent the nucleotide positions of the L-5′ UTR from the 3′ end of the virus-sense L segment. The 13-nt-long regions are contained in boxes, and the L mRNA transcriptional termination site is denoted by dotted lines with terminal arrows. The underlined nucleotides represent the mutated nucleotides that were introduced into LUTR-8, LUTR-9, LUTR-10, LUTR-11, LUTR-12, LUTR-13, LUTR-14, and LUTR-15. Northern blot analyses using intracellular RNAs from minigenome-replicating cells were performed using a probe that bound to the sense rLuc gene ORF (antisense). The anti-virus-sense S-like minigenome (Svc) and rLuc mRNA are shown. The efficiency of rLuc mRNA transcriptional termination of each minigenome (relative termination) was determined as described in the legend to Fig. 7. (B) The computer-predicted RNA secondary structures of the region from nt 6324 to 6389 of the nascent rLuc mRNAs of LUTR-1, LUTR-8, LUTR-9, LUTR-10, LUTR-11, LUTR-12, LUTR-13, LUTR-14, and LUTR-15, from nt 6324 to 6373 of that of LUTR-6, and from nt 6324 to 6368 of that of LUTR-7. The RNA secondary structure of the corresponding region of the nascent rLuc mRNA of rMP12-LUTR (see the legend to Fig. 4) is shown as LUTR (complementary). The most energetically stable structures at 37°C are shown. The RNA secondary analysis of LUTR-1 and its mutants used the anti-virus-sense RNA sequence, while that of LUTR (complementary) used the virus-sense RNA sequence. Nucleotide positions corresponding to LUTR-1 are shown in the structures. Boxes represent the L mRNA termination element, which consisted of nt 6366 to 6368 and nt 634 to 6345. Elements I and II are shown in boxes. The asterisks represent an RNA secondary structure that is similar to element I. dG, initial free energy (kcal/mol).