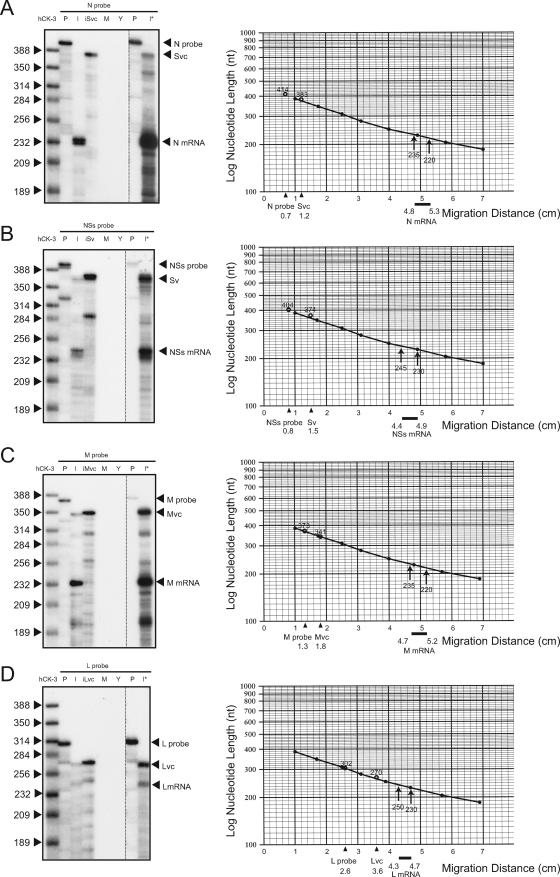
FIG. 2.
RPA of the 3′ termini of N, NSs, M, and L mRNAs. Total intracellular RNA from mock-infected Vero E6 cells (M) and that from MP-12-infected Vero E6 cells (I); yeast tRNA (Y); in vitro-transcribed, anti-virus-sense S RNA (iSvc); in vitro-transcribed, virus-sense S RNA (iSv); in vitro-transcribed, anti-virus-sense M RNA (iMvc); or in vitro-transcribed, anti-virus-sense L RNA (iLvc) were mixed with radiolabeled N probe (A), NSs probe (B), M probe (C), or L probe (D), and RPAs were performed (left panels). A commercially available multiprobe (hCK-3) served as a size marker. An undigested probe that was used for each RPA was also applied to the gel (P). Lanes I* used larger amounts of RNA from infected cells. Standard curves were plotted on semilog graphs based on the migration of the hCK-3 marker (right panels). The x axes show migrations (in centimeters) of the signals of the undigested probes, those of the viral genomic RNAs, and those of the viral mRNAs from the top of the gels.