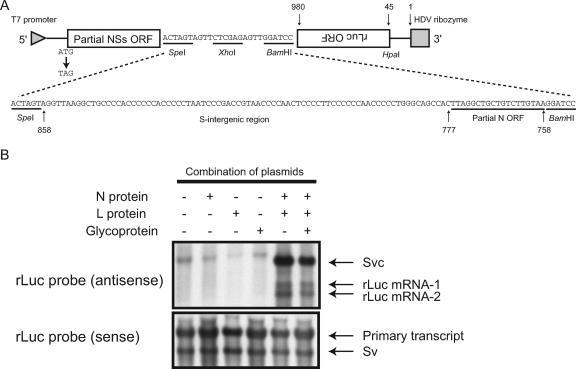
FIG. 6.
Analysis of transcriptional termination using a minigenome system. (A) Schematic diagram of pPro-T7-mSv expressing an S-segment-like minigenome (top). The sequence shown at the bottom represents the inserted S-IGR in IGR-1. The nucleotide positions of the pPro-T7-mSv from the 3′ end of the virus-sense S-like minigenome and those of the S-intergenic region and the partial N ORF from the 3′ end of the virus-sense S segment are shown at the top and bottom, respectively. (B) Subconfluent BHK/T7-9 cells in 6-well plates were transfected with 1.5 μg of IGR-1, 1.0 μg of pT7-IRES-vN (N protein), 0.5 μg of pT7-IRES-vL (L protein), or 1.0 μg of pCAGGS-vG (Glycoprotein). At 48 h posttransfection, intracellular RNAs were extracted and analyzed by Northern blotting using strand-specific probes that bound to the sense rLuc gene ORF (antisense) or the antisense rLuc gene ORF (sense). The transcript without the self cleavage is shown along with the HDV ribozyme (primary transcript), the anti-virus-sense IGR-1 (Svc), the virus-sense IGR-1 (Sv), and two rLuc mRNAs (rLuc mRNA-1 and rLuc mRNA-2).