Abstract
The interferon-stimulated genes (ISGs) ISG56 and ISG54 are strongly induced in cultured cells by type I interferons (IFNs), viruses, and double-stranded RNA (dsRNA), which activate their transcription by various signaling pathways. Here we studied the stimulus-dependent induction of both genes in vivo. dsRNA, which is generated during virus infection, induced the expression of both genes in all organs examined. Induction was not seen in STAT1-deficient mice, indicating that dsRNA-induced gene expression requires endogenous IFN. We further examined the regulation of these ISGs in several organs from mice injected with dsRNA or IFN-β. Both ISG56 and ISG54 were widely expressed and at comparable levels. However, in organs isolated from mice injected with IFN-α the expression of ISG54 was reduced and more restricted in distribution compared with the expression level and distribution of ISG56. When we began to study specific cell types, splenic B cells showed ISG54 but not ISG56 expression in response to all agonists. Finally, in livers isolated from mice infected with vesicular stomatitis virus, the expression of ISG56, but not ISG54, was induced; this difference was observed at both protein and mRNA levels. These studies have revealed unexpected complexity in IFN-stimulated gene induction in vivo. For the first time we showed that the two closely related genes are expressed in a tissue-specific and inducer-specific manner. Furthermore, our findings provide the first evidence of a differential pattern of expression of ISG54 and ISG56 genes by IFN-α and IFN-β.
The interferon (IFN) system is the first line of defense against virus infection in mammalian cells (15, 44). The antiviral effects of interferons are mediated by proteins encoded by IFN-stimulated genes (ISGs), whose transcription is induced by the Jak-STAT pathway (9, 10, 36, 49). The binding of type I IFNs, IFN-α and IFN-β, to their cell surface receptor (IFNAR) leads to Jak1- and Tyk2-mediated tyrosine phosphorylation of STAT1 and STAT2, which heterodimerize, bind to IFN regulatory factor 9 (IRF-9, or p48) to form the IFN-stimulated gene factor 3 (ISGF3) and translocate to the nucleus (8, 30). Once in the nucleus, ISGF3 binds to the interferon-stimulated response element (ISRE) present in the promoter regions of all ISGs and activates their transcription (7, 29, 37). IRF-9 is the component of ISGF3 which recognizes ISREs, and these elements can be recognized by other members of the IRF family, most notably IRF-3 and IRF-7 (16, 42, 45). As such, signaling pathways which lead to the activation of other IRFs can induce transcription of ISRE-containing genes without the involvement of IFNs. These viral stress-inducible genes (VSIGs) are induced by many viruses and other infectious agents, even in the absence of functional Jak-STAT signaling (46). Among the most highly induced VSIGs are the members of the ISG56 gene family. Four members of the family have been identified in humans, (ISG56/IFIT-1, ISG54/IFIT-2, ISG58/IFIT-5, and ISG60/IFIT-4), whereas in the mouse there are three members (ISG56/IFIT-1, ISG54/IFIT-2, and ISG49/IFIT-3) (5, 11, 29, 41, 57). These genes are phylogenetically related, clustered on the same chromosomes, and often coordinately induced in response to IFNs, dsRNA, or viral infection (12, 18, 27, 38, 48, 52, 56).
Several partially overlapping signaling pathways involved in antiviral defense can activate IRF-3 or IRF-7, resulting in the induction of VSIG transcription. Toll-like receptor 3 (TLR3) is a receptor for dsRNA located on endosome membranes (2). Downstream of TLR3 the adaptor protein TRIF recruits the protein kinase TBK-1, which phosphorylates IRF-3, causing its dimerization and nuclear translocation (31, 33). Complete activation of IRF-3 requires its additional phosphorylation by a phosphatidylinositol 3-kinase-mediated pathway (40). Viral single-stranded RNA can bind to TLR7 or TLR8 to activate similar pathways to TLR3 (19, 53). Viral CpG DNA or glycoproteins can trigger TLR9 or TLR4, respectively, to activate similar signaling pathways (20, 22). Signaling by all of these receptors converges on TBK-1 and IRF-3/IRF-7. Several cytoplasmic dsRNA-binding proteins, such as PKR, RIG-I, and Mda-5, have also been implicated in dsRNA-mediated and antiviral signaling (43, 58, 59). Among these alternative pathways, the ones initiated by the cytoplasmic RNA helicases RIG-I and Mda-5 appear most important for induction of IRFs (3, 14, 59). They use the adaptor protein IPS-1 to recruit TBK-1 and activate IRF-3 (25).
The most highly homologous proteins encoded by the ISG56 family members show only 70% sequence identity. However, they all contain multiple tetratricopeptide repeat motifs, which are degenerate protein interaction modules facilitating specific interactions with other cellular proteins (47). Human and mouse p56 and p54 inhibit initiation of translation by binding to various subunits of the translation initiation factor, eIF3, a large multisubunit protein complex with multiple functions in translation initiation (21, 34). Binding of these proteins to different subunits of eIF3 has diverse functional consequences. Human p56 and p54, both of which bind to the e subunit, block eIF3-mediated stabilization of the eIF2·GTP·Met-tRNA ternary complex (17, 23, 51). In contrast, mouse p56 and p54 and human p54, all of which bind to the c subunit of eIF3, block the ability of eIF3 to promote the formation of the 48S preinitiation complex containing the 40S ribosomal subunit, the ternary complex, eIF4F, and mRNA (24, 51, 52). The translation-inhibitory effect of human p56 has been suggested to be one of the major antiviral mechanisms used by IFNs to block the replication of hepatitis C virus (50).
Little is known about the regulation of expression of the ISG56 family of genes in vivo. Recently, Wacher et al. (55) examined their expression in the brains of mice infected with lymphocytic choriomeningitis virus or West Nile virus. Using in situ hybridization, they demonstrated that these genes are widely expressed in the neurons and other brain cells of virus-infected mice. In the current study, we have investigated the induction patterns of mouse p56 and p54 in vivo, in response to intravenous injection of dsRNA, IFN-α, IFN-β, or vesicular stomatitis virus (VSV). Newly developed antisera to the two proteins were used to examine various mouse tissues for the presence of the two proteins. Our study revealed an unexpected and interesting complexity of tissue-specific and inducer-specific noncoordinate regulation of expression of the two proteins.
MATERIALS AND METHODS
Mice.
Unless otherwise indicated, all experiments were performed with male FVB mice obtained from Taconic Farms. Age-matched wild-type (wt) and stat1−/− mice (129Sv background) were obtained from G. M. Chisolm (The Lerner Research Institute, Cleveland, OH). The wt control, tlr3−/−, pkr−/−, and tlr3−/− pkr−/− (double knockout [DKO]) mice were in a C57BL/6/129Sv mixed background. All mice were used at 8 to 10 weeks of age.
Injections.
Where indicated, mice were injected intravenously (i.v.) for 8 h with 2 × 105 U of recombinant murine IFN-α1, 100 μg dsRNA [poly(I)·poly(C); Pharmacia], 12 × 105 U rat IFN-β (generous gift from D. Lindner, The Lerner Research Institute, Cleveland, OH), or 4 × 106 PFU VSV (Indiana strain), a gift from A. Banerjee (The Lerner Research Institute, Cleveland, OH), each in 100 μl of phosphate-buffered saline (PBS). As controls, we also injected mice i.v. with 100 μl of PBS.
Antibodies.
Polyclonal antibodies to murine p56 and p54 were raised at the Hybridoma Core, Lerner Research Institute (Cleveland, OH), against the purified bacterially expressed full-length murine p56 or p54 proteins (52). Sera from injected rabbits were collected and tested for their anti-murine p56 or p54 activity by Western blotting.
Protein extraction and Western blotting.
Eight hours after i.v. injection, mice were sacrificed by CO2 inhalation and the organs were removed and immediately frozen in liquid nitrogen. Whole-cell protein extracts from the organs were made as previously described (32). Briefly, the organs were resuspended in 1 ml of lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, and protease inhibitor), homogenized using a Dounce homogenizer on ice, and then centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant was collected, and protein was measured. A 60-μg aliquot of extract was subjected to electrophoresis through a 10% denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. Western blotting was performed with a 1:2,000 dilution of a polyclonal anti-murine p56 antibody and 1:20,000 dilution of a polyclonal anti-murine p54 antibody.
All the experiments were done three times unless otherwise indicated in the figure legends. Since the results obtained showed similar induction patterns, representative experiments are shown in the figures.
RNase protection assay.
Frozen tissues were homogenized in a Dounce homogenizer on ice with 1 ml of TRIzol reagent (Invitrogen) and then RNA was isolated according to the manufacturer's protocol. RNase protection assays (RPAs) were performed with the RPA III kit (Ambion) following the manufacturer's protocol. The antisense probes to ISG56 and ISG54 were generated first by cutting the cDNAs with MboII and StuI, respectively, and then transcribing both with SP6 RNA polymerase. For each sample, 20 μg of total RNA was used for RPAs, and protected mRNA levels were visualized by autoradiography and quantified by a phosphorimager using Molecular Dynamics ImageQuant software (Amersham Bioscience). Levels of cyclophilin mRNA were used as an internal control.
Immunofluorescence and immunohistochemistry.
Mouse embryonic fibroblast (MEF) cells were grown on coverslips in six-well tissue culture plates and stimulated with or without 1,000 U/ml IFN-β for 16 h. Immunofluorescence assays were performed as described previously (35). Cells were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.2% Triton X-100 for 15 min. The cells were then blocked with PBS containing 0.02% Tween 20, 3% bovine serum albumin, and 3% goat serum at 4°C overnight and then incubated with primary antibody (1:1,500) for 2 h at room temperature. Cells were washed and further incubated with secondary antibody anti-rabbit-AlexaFluor 488 (1:1,500; Molecular Probes) for 1 h at room temperature. After incubation, cells were washed and covered with Vectashield containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Labs). Thereafter, the slides were resolved on a Leica digital fluorescent microscope.
For immunohistochemistry, the organs were Histochoice preserved and paraffin embedded, cross-sectioned, and deparaffined as described previously (26). Following paraffin removal, the slides were incubated in 10 mM sodium citrate pH 6 at 100°C for 10 min, and then in PBS for 5 min and in blocking solution at 4°C overnight. For antibody staining, the same conditions as reported above were followed except for the dilutions used for the primary antibody (1:1,000) and secondary antibody (anti-rabbit-AlexaFluor 488 from Molecular Probes, used at 1:200).
Flow cytometry.
Following harvest of spleens and bone marrow, single-cell suspensions were prepared by passing spleens through a 0.7-μm sieve (Falcon) and bone marrow through a 23-gauge needle. Following Tris-ammonium chloride lysis of red blood cells and washing with PBS, 106 cells per sample were stained. To stain splenocytes with cell surface markers, cells were incubated with an antibody to block Fc receptors (clone 2.4G2; BD Pharmingen) before addition of antibodies against CD4, CD8, or B220 (clones RM4-5, 53-6.7, and RA3-6B2, respectively; BD Pharmingen). Cells were washed twice with PBS before intracellular staining. For intracellular staining of splenocytes and bone marrow cells to study p54 and p56 induction, cells were fixed by incubation with 1% paraformaldehyde (Fisher Scientific). This was followed by washing with a fluorescence-activated cell sorter (FACS) buffer of PBS containing 2% fetal bovine serum (Atlanta Biologicals) and 10 mM sodium azide (Sigma). Cells were incubated with anti-murine p54 or p56 antibodies in FACS buffer plus 0.3% saponin (Sigma) before being washed twice in FACS buffer plus 0.03% saponin. Cells were then incubated with goat anti-rabbit immunoglobulin G conjugated to AlexaFluor 488 (Molecular Probes) followed by washing with FACS buffer plus 0.03% saponin and resuspended in FACS buffer for analysis. Flow cytometry was conducted on a FACScan flow cytometer (Becton Dickinson), and data were analyzed using FlowJo software (TreeStar Inc.).
Cells and cell transfection.
H1080 human fibrosarcoma cells (28) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin. Cells were transfected with Myc-pcDNA3 vector containing murine p56 or p54 or vector alone (52) using Fugene 6 (Boehringer Mannheim Co.). After 18 h, cells were collected and lysates were used for Western blotting.
RESULTS
Characterization of p54 and p56 antibodies.
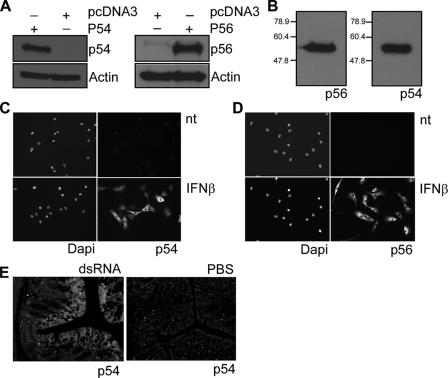
New polyclonal rabbit antibodies were raised against bacterially expressed, purified mouse p54 and p56. They specifically and efficiently recognized the cognate proteins in Western analysis (Fig. 1A). The two antisera were titrated to give equally strong signals with equal amounts of the two antigens; when equimolar amounts of the two recombinant proteins were analyzed, the anti-p54 serum was 10 times more potent than the anti-p56 serum (Fig. 1B). Both sera were assessed for use in immunofluorescence analyses (Fig. 1C and D); as expected, both proteins were absent in untreated MEFs and strongly induced by IFN-β, primarily in the cytoplasm. The p54 antiserum was also tested for use in immunohistochemistry. Mice were injected with dsRNA or PBS, and colonic sections were stained with anti-p54 serum (Fig. 1E). Low levels of p54 staining were present in the sample from the PBS-injected mouse (Fig. 1E, right), whereas strong signals were observed in the luminal epithelium in the mouse injected with dsRNA (Fig. 1E, left).
FIG. 1.
Characterization of anti-murine p56 and p54 polyclonal antibodies. (A) Antibodies raised against murine p54 and p56 proteins were tested in an immunoblot assay using extracts from HT1080 cells transfected with expression vector encoding mouse p54 or mouse p56 or with vector alone. (B) Purified proteins (50 ng) were immunoblotted with antibody against p56 (1:2,000) and p54 (1:20,000). (C and D) Intracellular localization of p54 and p56 proteins. MEF cells were treated with IFN-β for 16 h or PBS (nt), and then immunofluorescence with antibody to murine p54 or murine p56 was performed. Nuclei stained with DAPI are also shown. (E) Slides from the colons of mice injected with dsRNA or PBS for 8 h were immunostained with antibody against murine p54.
In vivo induction of p54 and p56 by VSV and dsRNA.
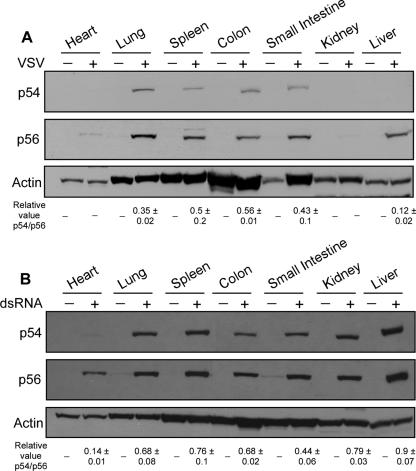
To examine the induction patterns of p54 and p56 in virus-infected mice, tissues were harvested 8 h after intravenous injection with VSV or PBS and subjected to Western blotting analysis. VSV induced p56 in all tissues except heart and kidney. Induction of p54 was less than p56 in all tissues. Strikingly, in the liver, p54 was hardly induced, although p56 was strongly induced (Fig. 2A).
FIG. 2.
Profiles of p54 and p56 protein expression following VSV and dsRNA injection. Mice were injected with PBS or VSV (A) or dsRNA (B), and after 8 h multiple organs were removed and protein extracts subjected to Western blotting with murine p56 and p54 polyclonal antibodies. The number at the bottom of each lane shows the mean and standard deviation of the relative p54/p56 value. Two independent experiments for panel A and three independent experiments for panel B were done.
To investigate if the expression patterns of p54 and p56 induced by VSV were mediated by dsRNA, a by-product generated by virus infection, we intravenously injected dsRNA or PBS alone in mice and performed Western blotting on tissue harvested 8 h postinjection. While neither protein was detected in most of the tissues analyzed from PBS-treated mice, we observed that a low level of p56, but not p54, was expressed in the small intestine. However, both proteins were strongly induced in most tissues of mice injected with dsRNA (Fig. 2B). A notable exception was the heart, in which induction of p56 but not p54 was observed.
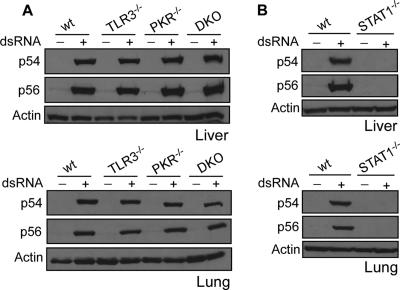
Genetically modified mice were used to investigate the potential role of TLR3 and PKR, two dsRNA-binding proteins, in mediating gene induction in response to dsRNA in vivo. Both p56 and p54 were strongly induced by dsRNA in the liver, the lung, and other tissues of both tlr3−/− and pkr−/− mice (Fig. 3A and data not shown). To investigate whether TLR3 and PKR can substitute for each other in mediating dsRNA-elicited signaling, a tlr3−/− pkr−/− DKO mouse strain was generated. Both proteins were induced in the DKO mouse as well, demonstrating conclusively that neither PKR nor TLR3 was required for gene induction in mice by intravenous injection of dsRNA (Fig. 3A).
FIG. 3.
Levels of dsRNA-induced p54 and p56 protein expression in tlr3−/−, pkr−/−, and stat1−/− mice. Eight hours after injection with PBS or dsRNA, livers and lungs were isolated from wt, tlr3−/−, pkr−/−, and tlr3−/− pkr−/− mice (A) or wt and stat1−/− mice (B). Extracts were prepared from the organs and used for Western blot assays with antibodies against murine p54 and p56.
To investigate the role of the IFN system in mediating in vivo gene induction by dsRNA, a different mouse strain was used. Because STAT1 is required for signal transduction by both type I and type II IFN, but not dsRNA, we chose stat1−/− mice to test whether gene induction was a direct result of dsRNA signaling or dependent on IFN produced in response to dsRNA. Neither protein could be induced by dsRNA in the liver, lung, or other tissues of stat1−/− mice (Fig. 3B and data not shown), indicating that at least in the tissues examined, IFN signaling was required for p54 and p56 induction by dsRNA.
Induction of p54 and p56 by type I IFNs in vivo.
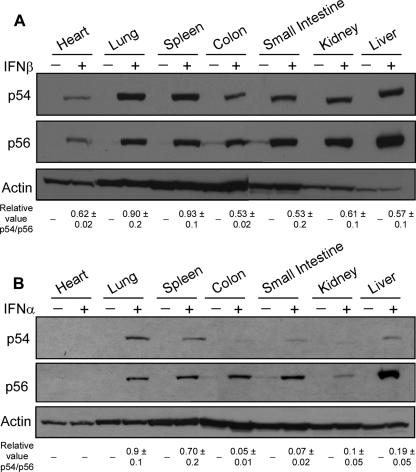
Induction of p54 and p56 by type I IFNs in different tissues of mice was investigated next. Because pure mouse IFN-β was not available in large quantities, we used rat IFN-β. In vitro testing of this IFN-β preparation revealed that it was about six times less active in mouse cells then in rat cells; we therefore used six times more rat IFN-β in vivo to compare its gene-inducing activity with that of mouse IFN-α. Interestingly, a low level of p56 but not p54 was expressed in the colon and small intestine, even in the absence of IFN treatment. IFN-β, injected intravenously, induced both proteins efficiently 8 h postinjection (Fig. 4A). In all tissues examined, both p54 and p56 were induced by IFN-β, although in some tissues, such as the liver, they were induced more strongly than in others, such as the heart, Surprisingly, injected mouse IFN-α produced different results (Fig. 4B). When we administered IFN-α by the same route as IFN-β, a variety of induction patterns were observed in different mouse tissues. In the heart, neither p54 nor p56 was induced by IFN-α, whereas in the lung and the spleen both were induced substantially. In contrast, in the colon, the small intestine, and the liver, p56 was induced very strongly but p54 was barely induced. In the kidney, p56 induction was barely detectable and p54 was not induced. These results demonstrated that different mouse tissues respond differently to intravenous IFN-β and IFN-α. Moreover, p54 and p56 are not always coordinately induced in all tissues.
FIG. 4.
Profiles of p54 and p56 protein expression following IFN-β and IFN-α injection. Mice were injected with PBS, IFN-β (A), or IFN-α (B), and after 8 h multiple organs were removed and protein was extracted. p54 and p56 protein levels were analyzed by Western blotting. Data are means ± standard deviations of relative values of the p54/p56 ratio after stimulation. Results shown are means of three independent experiments.
Induction of p56 and p56 in T and B cells.
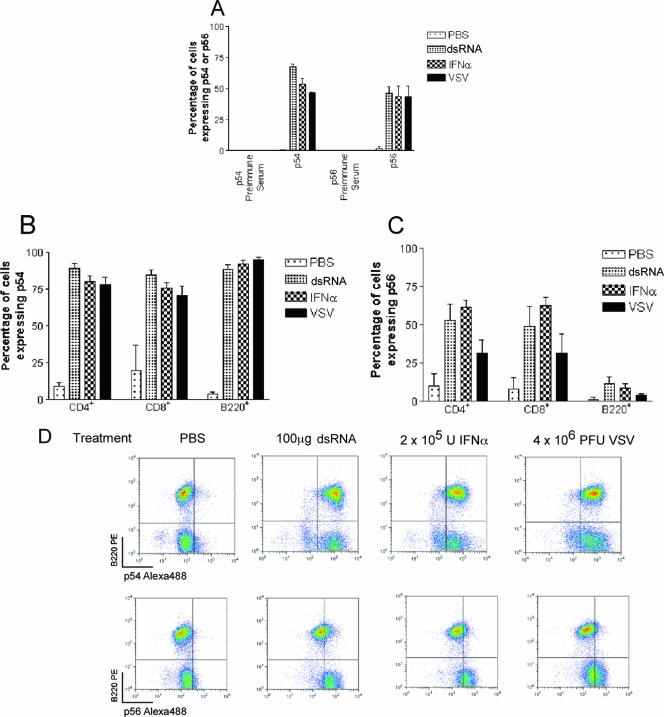
Induction of the p54 and p56 proteins in CD4+ and CD8+ T cells and B220+ B cells was examined by intracellular staining and flow cytometry. Cells were harvested for analysis 8 h after intravenous injection of dsRNA, IFN-α, or vesicular stomatitis virus. In bone marrow cells, all stimuli induced both p56 and p54, whereas cells from PBS-injected mice expressed very little of the two proteins (Fig. 5A). In T cells derived from the spleen, p54 was strongly stimulated by all three inducers (Fig. 5B). In contrast, p56 was less strongly induced, especially by VSV (Fig. 5C). On the other hand, in B cells derived from the spleen, p56 was scarcely induced by dsRNA, IFN-α, and VSV, whereas p54 was strongly stimulated by all inducers (Fig. 5C and D). These results revealed a clear discordance between the induction patters of p56 and p54 proteins in B cells.
FIG. 5.
Patterns of p54 and p56 protein expression in bone marrow and splenocytes upon injection of dsRNA, IFN-α, and VSV. Eight hours after i.v. injection of PBS, dsRNA, IFN-α, or VSV, cells from bone marrow and spleen were isolated and analyzed by flow cytometry. (A) p54 and p56 protein expression levels in bone marrow. Flow cytometry data were analyzed as for panel D, and the percentage of cells showing induction of p54 or p56 was determined. (B and C) p54 (B) and p56 (C) protein expression in CD4+ T cells, CD8+ T cells, and B220+ B cells from spleens of mice treated as for panel A. Whole spleen preparations were stained with antibodies to CD4, CD8, or B220 followed by intracellular staining for p54 and p56 as described in Materials and Methods. Flow cytometry data were analyzed as for panel D, and the proportion of cells which induced p54 or p56 in gated CD4+, CD8+, or B220+ populations was determined. The CD4+ population contained at least 5,000 cells, the CD8+ population contained at least 2,000 cells, and the B220+ population contained at least 10,000 cells, in line with the proportions of these cell types in the spleen. (D) Induction of p54 but not p56 in response to stimuli in B220+ cells. Quadrant gates were set using preimmune serum controls to determine background levels of staining, and the percentage of lymphocytes in each quadrant is shown. Panels A to C show means ± standard deviations of three experiments. Density plots in panel D are representative of three experiments.
Differential inducer- and tissue-specific induction of ISG54 and ISG56 mRNAs.
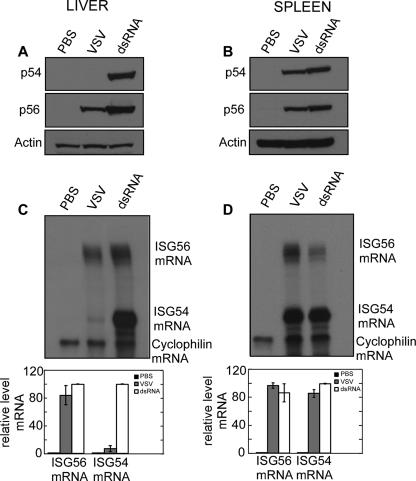
To document the characteristics of differential induction patterns of the two proteins and determine the molecular basis, we focused on two tissues, liver and spleen. In the spleen both VSV and dsRNA induced p54 and p56 proteins at comparable levels (Fig. 6B). On the contrary, in the liver dsRNA induced both proteins to a similar extent while VSV induced only p56 (Fig. 6A). The same patterns were reflected at the mRNA level. The levels of ISG54 mRNA and ISG56 mRNA were quantified by RNase protection assays using the cyclophilin mRNA level as the internal control. In the spleen, both mRNAs were induced by both stimuli (Fig. 6D), whereas in the liver, VSV induced only ISG56 mRNA (Fig. 6C). These results demonstrated that differential induction of the two ISGs is regulated at the mRNA level.
FIG. 6.
Differential expression of p54 and p56 proteins upon VSV and dsRNA injection. Livers and spleens were isolated 8 h after injection of dsRNA or VSV. (A and B) Proteins were extracted, and expression of p54 and p56 proteins was detected by Western blotting. (C and D) RNA was extracted, and ISG54 and ISG56 mRNAs were measured by RPA. Cyclophilin mRNA levels were used as internal controls (upper panels). Protected mRNA levels were quantified by phoshorimager (lower panels). The values are means ± standard deviations of two independent experiments.
DISCUSSION
An antiserum previously generated against a p56 peptide (52) also interacted with several other cellular proteins and therefore was not useful for immunofluorescence or histochemical analyses of tissues. In contrast, the new antisera were produced against purified recombinant p56 and p54 and could be used at high dilutions with little nonspecific signal. The anti-p54 serum was more potent and, at a 10-fold higher dilution, it produced a signal similar to that of the anti-p56 serum (Fig. 1B). As expected, neither antiserum produced any immunofluorescence signal in untreated cells (Fig. 1C and D), confirming their high specificity. The p54 antiserum also gave a specific signal when used in immunohistochemistry (Fig. 1E). The development of these reagents will greatly facilitate biochemical studies of p54 and p56.
The two antisera were extensively used for Western blot analyses of tissues isolated from mice treated with various inducers of antiviral and type I IFN signaling. Unlike VSV, dsRNA potently induced these proteins in many tissues (Fig. 2B). Surprisingly, neither TLR3 nor PKR was required for their in vivo induction (Fig. 3A), most probably because the injected dsRNA was internalized in cells and detected by RIG-I or Mda5 to trigger the signaling pathways that led to p54 and p56 induction. Moreover, we observed no induction of the two proteins in stat1−/− mice (Fig. 3B), indicating that IFN signaling was required for their induction by dsRNA in vivo. This observation is consistent with those of Wacher et al. (55), who noted blunted and cell-restricted induction of these genes in brains from lymphocytic choriomeningitis virus-infected stat1−/− mice. Combined interpretation of the results shown in Fig. 3 suggests that in some cells, possibly dendritic cells or macrophages which have high endocytic capacity, injected dsRNA is recognized by the cytoplasmic RIG-I/Mda-5 signaling systems and causes rapid production of IFN, which circulates and induces p54 and p56 in the majority of tissues. In the absence of IFN signaling in the stat1−/− mouse, this secondary induction arm was eliminated.
Consistent with the above model, direct injection of IFN-β, the main class of IFN induced by poly(I)·poly(C) (4), stimulated the expression of both proteins in all tissues examined (Fig. 4A). However, the magnitude of induction was variable, being the strongest in the liver and the weakest in the heart. This variability may result from differences in the bioavailability of injected IFN-β to different tissues. Tissue response differences were more prominent in mice injected with IFN-α (Fig. 4B). Moreover, there was a strong discordance between p54 and p56 induction levels in many tissues. In the lung and spleen, both proteins were induced to similar levels, whereas in several other tissues, including the liver, p56 was induced much more strongly than p54. A similar tissue-specific differential induction of the two proteins was observed in mice injected with VSV, which induces primarily IFN-α (Fig. 2A and 6). In the spleen, both proteins were induced by VSV and dsRNA, but in the liver, VSV induced only p56. This difference was reflected at the mRNA level, indicating that differential protein synthesis or turnover was not responsible for the phenomenon (Fig. 6).
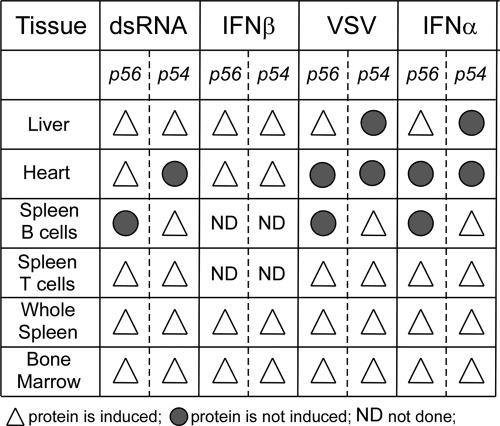
The unexpected and interesting differences in the tissue-specific gene induction patterns are summarized in Fig. 7. Bone marrow, whole spleen, spleen T cells (Fig. 7, bottom three lines), and many other tissues qualitatively behaved as expected; both p56 and p54 were induced by all four inducers. In contrast, this concordance was not seen in several other tissues. The liver, the heart, and B cells were useful examples of different types of discordance. In B cells, p54 was efficiently induced in response to all stimuli, whereas we could not detect p56 protein expression in response to any inducer tested. In the heart, dsRNA could induce p56 but not p54, and in spite of the fact that IFN-β could induce both genes, neither of them was expressed when we injected IFN-α. In the liver, the situation was even more complex: poly(I)·poly(C) and IFN-β induced both proteins, whereas VSV and IFN-α induced only p56. Although these differences were not absolute, they were quantitatively highly significant. Our results are strongly supportive of the notion that signaling by all type I IFNs in various tissues or cells does not occur by identical pathways.
FIG. 7.
Summary of differential induction of p54 and p56 proteins.
Three major mechanistic questions arise from these observations: how does a tissue such as the colon respond differently to related stimuli, such as IFN-α and IFN-β? How and why do tissues such as the spleen and kidney respond differently to IFN-α? Finally, how are two closely related genes, ISG56 and ISG54, differentially induced by the same stimulus in a given tissue, such as the liver or B cells? Published reports have shown that genes such as β-R1 (39) and CXCLII are selectively induced by IFN-β, but not IFN-α, in cell cultures (6). Here we report, for the first time, that in vivo another gene, ISG54, is preferentially induced by IFN-β compared to IFN-α in the liver. The primary Jak-STAT pathway cannot explain entirely the biological response to type I IFN; cell-specific and IFN class-specific ancillary signaling pathways may contribute to the ultimate constellations of genes induced. Different responses to IFN-α and IFN-β may also result from variations in stability of signaling complexes downstream of these inducers that are modulated by proteins such as TYK2 (13), variations in STAT activation patterns in response to these two stimuli (54), or alternative tyrosine phosphorylation patterns of the common IFNAR (1). Alternatively, the difference may not be at the transcriptional level but instead at the level of mRNA stability. The p54 mRNA may be inherently much less stable than p56 mRNA, and IFN-β, but not IFN-α, may provide an additional p54 mRNA stabilizing signal. These mechanisms for creating different responses to IFN-α and IFN-β within a given tissue may also be responsible for the different responses to a particular stimulus seen in various tissues. However, these mechanisms do not explain why both p54 and p56 are induced in some tissues but not others. It is possible that requirements for p54 and p56 vary between tissues depending on factors such as the protein turnover rate, accessibility to viral infection, or sensitivity to apoptotic death of cells in the organ as a consequence of translation inhibition. Finally, how would ISG54 and ISG56 behave differently in some tissues? Although the two genes are clustered on the same mouse chromosome and have the same promoter elements (5), it is tempting to speculate that in B cells, ISG56 is not accessible to transcription factors because of its location in a silent region of the chromatin and therefore cannot be induced. It remains to be seen whether the same is true for B cells infected with other viruses or treated with IFNs ex vivo. For productively pursuing the above mechanistic questions, we need to reproduce the critical phenomena in cell culture models. Only in this way can appropriate analytical experiments be executed. The in vivo study reported here has provided us with interesting observations that raise important questions, but the complexity of the in vivo system is not conducive to the necessary mechanistic experiments regarding regulation of gene expression. The current study, however, has highlighted several new physiological aspects of the IFN system, namely, that IFN-α and IFN-β are not equivalent, two closely related ISGs are not always regulated similarly, and different cell types respond quite differently to different inducers.
Acknowledgments
We thank G.M. Chisolm for the kind gift of wt and stat1−/− mice, D. Lindner for the kind gift of rat IFN-β, A. Banerjee for the gift of VSV, and Earl Poptic from the Lerner Research Institute Hybridoma Core for producing the antibodies.
This work was supported by National Institutes of Health grants CA068782 and CA062220.
Footnotes
Published ahead of print on 6 June 2007.
REFERENCES
- 1.Abramovich, C., L. M. Shulman, E. Ratovitski, S. Harroch, M. Tovey, P. Eid, and M. Revel. 1994. Differential tyrosine phosphorylation of the IFNAR chain of the type I interferon receptor and of an associated surface protein in response to IFN-alpha and IFN-beta. EMBO J. 13:5871-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 3.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barchet, W., M. Cella, B. Odermatt, C. Asselin-Paturel, M. Colonna, and U. Kalinke. 2002. Virus-induced interferon alpha production by a dendritic cell subset in the absence of feedback signaling in vivo. J. Exp. Med. 195:507-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bluyssen, H. A., R. J. Vlietstra, P. W. Faber, E. M. Smit, A. Hagemeijer, and J. Trapman. 1994. Structure, chromosome localization, and regulation of expression of the interferon-regulated mouse Ifi54/Ifi56 gene family. Genomics 24:137-148. [DOI] [PubMed] [Google Scholar]
- 6.Coelho, L. F., G. Magno de Freitas Almeida, F. J. Mennechet, A. Blangy, and G. Uze. 2005. Interferon-alpha and -beta differentially regulate osteoclastogenesis: role of differential induction of chemokine CXCL11 expression. Proc. Natl. Acad. Sci. USA 102:11917-11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, B., D. Peretz, D. Vaiman, P. Benech, and J. Chebath. 1988. Enhancer-like interferon responsive sequences of the human and murine (2′-5′) oligoadenylate synthetase gene promoters. EMBO J. 7:1411-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 9.Decker, T., S. Stockinger, M. Karaghiosoff, M. Muller, and P. Kovarik. 2002. IFNs and STATs in innate immunity to microorganisms. J. Clin. Investig. 109:1271-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Veer, M. J., H. Sim, J. C. Whisstock, R. J. Devenish, and S. J. Ralph. 1998. IFI60/ISG60/IFIT4, a new member of the human IFI54/IFIT2 family of interferon-stimulated genes. Genomics 54:267-277. [DOI] [PubMed] [Google Scholar]
- 12.Elco, C. P., J. M. Guenther, B. R. Williams, and G. C. Sen. 2005. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-κB, and interferon but not toll-like receptor 3. J. Virol. 79:3920-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamero, A. M., R. Potla, J. Wegrzyn, M. Szelag, A. E. Edling, K. Shimoda, D. C. Link, J. Dulak, D. P. Baker, Y. Tanabe, J. M. Grayson, and A. C. Larner. 2006. Activation of Tyk2 and Stat3 is required for the apoptotic actions of interferon-beta in primary pro-B cells. J. Biol. Chem. 281:16238-16244. [DOI] [PubMed] [Google Scholar]
- 14.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 103:8459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 16.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19:6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267:209-219. [DOI] [PubMed] [Google Scholar]
- 19.Heil, F., H. Hemmi, H. Hochrein, F. Ampenberger, C. Kirschning, S. Akira, G. Lipford, H. Wagner, and S. Bauer. 2004. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303:1526-1529. [DOI] [PubMed] [Google Scholar]
- 20.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 21.Hershey, J. W., and W. C. Merrick. 2000. Pathway and mechanism of initiation of protein synthesis, p. 1-55. In J. W. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control of gene expression. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 22.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Cutting edge. Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162:3749-3752. [PubMed] [Google Scholar]
- 23.Hui, D. J., C. R. Bhasker, W. C. Merrick, and G. C. Sen. 2003. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J. Biol. Chem. 278:39477-39482. [DOI] [PubMed] [Google Scholar]
- 24.Hui, D. J., F. Terenzi, W. C. Merrick, and G. C. Sen. 2005. Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. J. Biol. Chem. 280:3433-3440. [DOI] [PubMed] [Google Scholar]
- 25.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 26.Kessler, S. P., P. S. Senanayake, T. S. Scheidemantel, J. B. Gomos, T. M. Rowe, and G. C. Sen. 2003. Maintenance of normal blood pressure and renal functions are independent effects of angiotensin-converting enzyme. J. Biol. Chem. 278:21105-21112. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura, Y., O. Spleiss, H. Li, T. Taniguchi, H. Kimura, Y. Nomura, and P. J. Gebicke-Haerter. 2001. Lipopolysaccharide-induced switch between retinoid receptor (RXR) alpha and glucocorticoid attenuated response gene (GARG)-16 messenger RNAs in cultured rat microglia. J. Neurosci. Res. 64:553-563. [DOI] [PubMed] [Google Scholar]
- 28.Leonard, G. T., and G. C. Sen. 1997. Restoration of interferon responses of adenovirus E1A-expressing HT1080 cell lines by overexpression of p48 protein. J. Virol. 71:5095-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy, D., A. Larner, A. Chaudhuri, L. E. Babiss, and J. E. Darnell, Jr. 1986. Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc. Natl. Acad. Sci. USA 83:8929-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy, D. E., and J. E. Darnell, Jr. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3:651-662. [DOI] [PubMed] [Google Scholar]
- 31.McWhirter, S. M., K. A. Fitzgerald, J. Rosains, D. C. Rowe, D. T. Golenbock, and T. Maniatis. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. USA 101:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meraz, M. A., J. M. White, K. C. Sheehan, E. A. Bach, S. J. Rodig, A. S. Dighe, D. H. Kaplan, J. K. Riley, A. C. Greenlund, D. Campbell, K. Carver-Moore, R. N. DuBois, R. Clark, M. Aguet, and R. D. Schreiber. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431-442. [DOI] [PubMed] [Google Scholar]
- 33.Oganesyan, G., S. K. Saha, B. Guo, J. Q. He, A. Shahangian, B. Zarnegar, A. Perry, and G. Cheng. 2006. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439:208-211. [DOI] [PubMed] [Google Scholar]
- 34.Pain, V. M. 1996. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 236:747-771. [DOI] [PubMed] [Google Scholar]
- 35.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NFκB- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. USA 99:6322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 37.Porter, A. C., Y. Chernajovsky, T. C. Dale, C. S. Gilbert, G. R. Stark, and I. M. Kerr. 1988. Interferon response element of the human gene 6-16. EMBO J. 7:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rani, M. R., G. R. Foster, S. Leung, D. Leaman, G. R. Stark, and R. M. Ransohoff. 1996. Characterization of beta-R1, a gene that is selectively induced by interferon beta (IFN-beta) compared with IFN-alpha. J. Biol. Chem. 271:22878-22884. [DOI] [PubMed] [Google Scholar]
- 40.Sarkar, S. N., K. L. Peters, C. P. Elco, S. Sakamoto, S. Pal, and G. C. Sen. 2004. Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat. Struct. Mol. Biol. 11:1060-1067. [DOI] [PubMed] [Google Scholar]
- 41.Sarkar, S. N., and G. C. Sen. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol. Ther. 103:245-259. [DOI] [PubMed] [Google Scholar]
- 42.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 43.Saunders, L. R., and G. N. Barber. 2003. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 17:961-983. [DOI] [PubMed] [Google Scholar]
- 44.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 45.Servant, M. J., N. Grandvaux, and J. Hiscott. 2002. Multiple signaling pathways leading to the activation of interferon regulatory factor 3. Biochem. Pharmacol. 64:985-992. [DOI] [PubMed] [Google Scholar]
- 46.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 47.Sikorski, R. S., M. S. Boguski, M. Goebl, and P. Hieter. 1990. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60:307-317. [DOI] [PubMed] [Google Scholar]
- 48.Smith, J. B., and H. R. Herschman. 1996. The glucocorticoid attenuated response genes GARG-16, GARG-39, and GARG-49/IRG2 encode inducible proteins containing multiple tetratricopeptide repeat domains. Arch. Biochem. Biophys. 330:290-300. [DOI] [PubMed] [Google Scholar]
- 49.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 50.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terenzi, F., D. J. Hui, W. C. Merrick, and G. C. Sen. 2006. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 281:34064-34071. [DOI] [PubMed] [Google Scholar]
- 52.Terenzi, F., S. Pal, and G. C. Sen. 2005. Induction and mode of action of the viral stress-inducible murine proteins, P56 and P54. Virology 340:116-124. [DOI] [PubMed] [Google Scholar]
- 53.Uematsu, S., S. Sato, M. Yamamoto, T. Hirotani, H. Kato, F. Takeshita, M. Matsuda, C. Coban, K. J. Ishii, T. Kawai, O. Takeuchi, and S. Akira. 2005. Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-α induction. J. Exp. Med. 201:915-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Boxel-Dezaire, A. H., M. R. Rani, and G. R. Stark. 2006. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 25:361-372. [DOI] [PubMed] [Google Scholar]
- 55.Wacher, C., M. Muller, M. J. Hofer, D. R. Getts, R. Zabaras, S. S. Ousman, F. Terenzi, G. C. Sen, N. J. King, and I. L. Campbell. 2007. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J. Virol. 81:860-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, J., and I. L. Campbell. 2005. Innate STAT1-dependent genomic response of neurons to the antiviral cytokine alpha interferon. J. Virol. 79:8295-8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wathelet, M., S. Moutschen, P. Defilippi, A. Cravador, M. Collet, G. Huez, and J. Content. 1986. Molecular cloning, full-length sequence and preliminary characterization of a 56-kDa protein induced by human interferons. Eur. J. Biochem. 155:11-17. [DOI] [PubMed] [Google Scholar]
- 58.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 59.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]