Abstract
Primary differentiated respiratory epithelial cell cultures closely model the in vivo environment and allow for studies of innate immune responses generated specifically by epithelial cells, the primary cell type infected by human influenza A virus strains. We used primary murine tracheal epithelial cell (mTEC) cultures to investigate antiviral and cytokine responses to influenza A virus infection, focusing on the contribution of the RNA binding domain of the NS1 protein. rWSN NS1 R38A replication is attenuated in mTEC cultures; however, viral antigen is detected predominantly in ciliated cells, similar to wild-type virus. NS1 and NS1 R38A proteins display a primarily cytoplasmic localization in infected mTEC cultures. Increased production of tumor necrosis factor alpha, interleukin-6, and beta interferon is observed during rWSN NS1 R38A infection, and cytokines are secreted in a directional manner. Cytokine pretreatment of mTEC cultures and Vero cells suggest that rWSN NS1 R38A is more sensitive to the presence of antiviral/inflammatory cytokines than wild-type virus. Our results demonstrate that the RNA binding domain is a critical regulator of both cytokine production and cytokine sensitivity during influenza A virus infection of primary tracheal epithelial cells.
Influenza A virus is a significant cause of morbidity and mortality in human and animal populations each year (2, 12, 14, 43, 127). The virus is in the Orthomyxoviridae family, and its genome consists of eight single-stranded, negative-sense RNA segments that, depending on the virus strain, encode 10 or 11 major proteins. The primary site of replication during influenza A virus infection of humans is the epithelia of the respiratory tract (133).
Primary lung and airway epithelial cell cultures have been used to investigate cellular responses to infection with many respiratory viruses, including influenza A virus (3, 7, 11, 20, 39, 42, 49, 64, 65, 78, 106, 117, 120, 129), respiratory syncytial virus (RSV) (69, 135), hantavirus (95), human coronaviruses (104, 105, 121), human parainfluenza virus type 3 (134), and adenovirus (85, 86, 110, 131, 132). Unlike conventional transformed cell lines, primary airway cultures, when differentiated fully, can mirror the in vivo tissue by being organized into a multilayered, polarized culture consisting of multiple cell types (7, 42, 85, 110, 117, 134, 135).
Studies with influenza A virus strains and human primary airway cultures have characterized the distribution of virus receptors (α-2,3 and α-2,6 sialic acid), cell tropism of both human and avian virus strains (42, 49, 65, 106, 117), virus-induced cytokine secretion (3, 7, 11, 120), and the importance of neuraminidase for virus entry (64). The laboratory mouse model for influenza A virus infection has been used extensively (58, 80), and primary murine tracheal epithelial cell (mTEC) cultures support influenza A virus (42) replication and provide the opportunity to study the epithelial cell-specific innate immune responses to virus infection.
Cytokine production during influenza A virus infection is believed to be a key mediator of pathology and disease severity (46) and is especially important in infections with avian H5N1 viruses, where heightened cytokine production is thought to be responsible in part for the increased mortality observed in patients (9, 13, 83, 111). The viral nonstructural protein 1 (NS1) is known to be an important regulator of innate and adaptive immunity (4, 22, 36, 72, 73, 81, 113, 114) and inhibits host immune responses through two functional domains: an N-terminal RNA binding domain (88) and a C-terminal effector domain (53). The RNA binding domain consists of a six-helix bundle that, when present in a homodimer (10, 57, 76), can interact with various forms of RNA (35, 59, 76, 88-90, 122) through electrostatic interactions of basic amino acid residues (123). Mutational studies within the RNA binding domain demonstrated that the arginine at amino acid position 38 in NS1 is absolutely critical for RNA binding (123). However, this amino acid may also constitute part of a nuclear localization signal (NLS) in some virus strains (31, 53, 68, 73). The effector domain interacts with proteins involved in 3′ end cellular mRNA processing, inhibits mRNA export and pre-mRNA splicing of host cell transcripts (1, 8, 24, 59, 75, 79, 87, 89, 90), and interacts with components of the nuclear pore complex as well as the mRNA export machinery (97). Finally, the NS1 protein inhibits the activation and/or signaling of antiviral proteins, such as retinoic acid-induced gene I product (RIG-I) (32, 72, 81), protein kinase R (PKR) (4, 36, 52, 114), 2′,5′ oligoadenylate synthetase/RNase L (73), activators of mitogen-activated protein kinase (18, 33, 103), and transcription factors involved in type I interferon and inflammatory cytokine signaling (60, 113, 125). While NS1 combats the host cell innate immune response at many levels, strain-specific functions may exist (38, 43, 79, 113, 125). It was recently demonstrated that there are differential requirements for the NS1 N- and C-terminal domains among certain strains, and this in part determines the specific antiviral molecules/interferon pathway(s) affected most profoundly by each virus (48).
We are using differentiated, primary mTEC (130) cultures to investigate epithelial cell-specific cytokine responses to influenza A virus infection, specifically focusing on the contribution of the NS1 R38A mutation in regulating host innate immune responses. We demonstrate that rWSN NS1 R38A infection of mTEC cultures leads to increased production of beta interferon (IFN-β) and the inflammatory cytokines tumor necrosis factor α (TNF-α) and interleukin-6 (IL-6). Furthermore, rWSN NS1 R38A replication is sensitive to cytokine pretreatment, suggesting a critical role for NS1 R38 in the regulation of both cytokine production and cytokine sensitivity during influenza A virus infection.
MATERIALS AND METHODS
Reagents and antibodies.
The components in TEC basic medium (TEC basic), proliferation medium (TEC plus), and maintenance medium (TEC MM) have been described previously (94, 130). The mouse anti-NS1 antibody (hybridoma 1A7CL; 1:100 immunofluorescence) was a kind gift of Robert Webster (6). Other primary antibodies purchased and used were mouse anti-β tubulin IV (1:100 immunofluorescence; BioGenex, San Ramon, CA), goat anti-H1 hemagglutinin (HA-H0 subtype) A/PR8/34 sera (1:500 immunofluorescence; NIH/NIAD reference reagent V314-511-157), sheep antiserum to mouse IFN-α/β (NIH/NIAD reference reagent G024-501-568), control sheep sera (NIH/NIAD reference reagent G025-501-568), recombinant mouse IFN-β (1 × 106 U/ml; PBL Biomedical Laboratories, Piscataway, NJ), recombinant human IFN-β (1 × 106 U/ml; PBL Biomedical Laboratories). Secondary antibodies used in this study for immunofluorescence were donkey anti-mouse Alexa Fluor 488 (1:500) and anti-nuclear stain TO-PRO-3, purchased from Molecular Probes (Eugene, OR). The following antibodies were purchased from Jackson ImmunoResearch (West Grove, PA): goat anti-mouse conjugated to fluorescein isothiocyanate (1:250) and donkey anti-goat conjugated to rhodamine (1:250).
Cell lines.
Madin-Darby canine kidney cells (MDCK; American Type Culture Collection, Manassas, VA), Vero cells (Vero; American Type Culture Collection), and L929 murine fibroblasts (kindly provided by Michael Diamond, Washington University, St. Louis, MO) were cultured in Dulbecco's modified Eagle medium (DMEM; Sigma, St. Louis, MO) containing 10% fetal bovine serum (Atlanta Biologics, Atlanta, GA) with 100 U/ml of penicillin and 100 μg/ml of streptomycin (Invitrogen, Grand Island, NY) and maintained at 37°C in a humidified environment containing 5% CO2.
Antibody collection for anti-NS1 1A7CL was conducted as previously described (67).
Viruses.
Recombinant influenza viruses in the A/WSN/33 genetic background (rWSN) were generated using the 12-plasmid rescue system in 293T cells as described previously (77). Nucleotide changes (CGA at nucleotides 138 to 140 mutated to GCG) were introduced into the NS segment coding region of WSN (GenBank sequence M12597) by two-step PCR mutagenesis. The same mutation was introduced into the NS segment coding region of A/Udorn/73 (112). The absence of other nucleotide changes in the NS coding region was confirmed by DNA sequencing. Virus stocks were generated, and infectious virus was quantified by plaque assay using MDCK cells as previously described (66, 82).
Vesicular stomatitis virus (VSV) with an additional transcription unit for green fluorescent protein (GFP) expression (VSV-GFP) was a kind gift from John Rose (5).
mTEC cultures and virus infection.
The isolation, culture, and differentiation of mTEC cultures were performed as previously described (94, 130) by using 5- to 10-week-old female BALB/c mice. When the cultures reached a transepithelial resistance of more than 1,000 Ω·cm2, the apical medium was removed to create an air-liquid interface and the cultures were allowed to differentiate for 10 to 14 days before virus infection.
mTEC cultures were infected via the apical chamber with 3,600 PFU of virus diluted in warm DMEM containing penicillin-streptomycin in a total volume of 100 μl. If all cells in the culture were susceptible to influenza virus infection, this would correspond to a multiplicity of infection (MOI) of approximately 0.01. Since the cultures are pseudostratified, not all cells in the culture will be exposed to the apically administered virus inoculum, nor are all the cells in the culture susceptible to infection (42). The cells were incubated with virus at 37°C for 1 h, the inoculum was removed, and cells were washed three times with 200 μl of DMEM containing penicillin-streptomycin. After washing, 100 μl of DMEM containing penicillin-streptomycin and 500 μl of TEC MM (94) was placed in the apical and basolateral chambers, respectively. Apical and basolateral supernatants were collected at the indicated times postinfection and stored at −70°C.
Naïve mTEC cultures were pretreated via the basolateral chamber with 500 μl of basolateral supernatant from rWSN NS1 R38A-infected mTEC cultures at day 3 postinfection. After overnight pretreatment, basolateral chambers were washed twice with 500 μl DMEM containing penicillin-streptomycin and replaced with fresh TEC MM (94). The mTEC cultures were infected via the apical chamber with 3,600 PFU as described above. Apical and basolateral supernatants were collected at the indicated times postinfection and stored at −70°C. Virus was not detected in the basolateral supernatants used for pretreatment.
Indirect immunofluorescence confocal microscopy.
At the indicated times postinfection, mTEC cultures or MDCKs were washed three times with phosphate-buffered saline (PBS; GIBCO, Inc., Carlsbad, CA) and fixed in PBS containing 2% paraformaldehyde for 15 min at room temperature. Cells were washed three times and permeabilized with PBS containing 0.2% Triton X-100 and 0.1% sodium citrate for 10 min at room temperature. Cells were washed with PBS and incubated in PBS containing 3% normal goat or normal donkey serum and 0.5% bovine serum albumin (blocking buffer) for 30 min at room temperature. Cells were washed again, incubated with blocking buffer containing primary antibodies for 1 h at room temperature, washed again, and incubated with blocking buffer containing secondary antibodies for 45 min. Where indicated, the nuclear stain TO-PRO-3 (Molecular Probes, Invitrogen) was included in the secondary antibody incubation. The wash solution for all steps is PBS with 0.2% Tween 20. Transwell-Clear membranes were mounted using 10 μl of Molecular Probes ProLong antifade (Molecular Probes, Invitrogen), and slides were imaged using a Zeiss LSM 510 Meta confocal microscope. Images for Fig. 3 were obtained with a 63× oil objective, and shown are flattened reconstructions from a 10-μΜ Z stack with 0.6-μM slices. Images for Fig. 4 were obtained with a 100× oil objective with a 2.5× digital zoom. NS1 images in the top two panels of Fig. 4 are single-slice images from an 8-μM Z stack with 0.6-μM slices. The filamentous NS1 image (Fig. 4, lower panels) shown is a flattened reconstitution from a 10-μM Z stack with 0.6-μM slices.
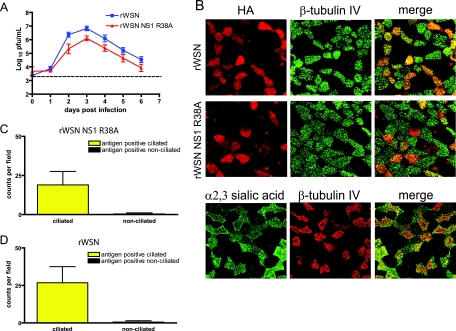
FIG. 3.
rWSN NS1 R38A replication, antigen expression, and tropism in mTEC cultures. (A) Replication of rWSN and rWSN NS1 R38A in mTECs. Cells were infected at days 10 to 14 after the air-liquid interface with a low MOI (3,600 PFU) and were monitored for the presence of virus in apical and basolateral chambers. Shown are the apical chamber virus titers. Virus was not detected in the basolateral supernatants at any time point. Data points are the averages of four separate experiments, and error bars indicate standard errors of the means. (B) rWSN and rWSN NS1 R38A antigen expression (upper panels) at day 3 postinfection. Red, HA; green, β-tubulin IV. (Lower panel) Expression of α2,3-linked sialic acid (MAA lectin, green) and β-tubulin IV (red) in mTEC cultures. All images were taken at ×63 magnification and are reconstructed from serial optical sections. (C and D) Quantitation of antigen-positive cells for rWSN and rWSN NS1 R38A. Cells in 10 to 15 fields were counted in three separate experiments. The data points are average values with standard errors of the means (error bars).
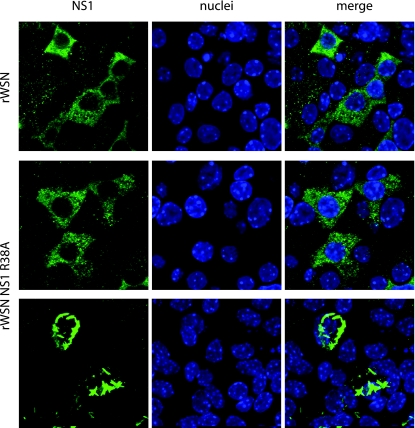
FIG. 4.
NS1 subcellular localization in infected mTEC cultures. NS1 subcellular localization in mTEC cultures at day 3 postinfection. Cells were immunostained with anti-NS1 antibody and the nuclear counterstain TO-PRO-3. (Upper panels) NS1 expression in rWSN-infected cells. (Middle panels) NS1 expression in the majority of rWSN NS1 R38A-infected cells. (Lower panels) Filamentous structures present in a minority of cells infected with rWSN NS1 R38A. All images were taken at ×100 magnification with a 2.5× digital zoom. NS1 images in the top two panels are single-slice images from an 8-μM Z stack with 0.6-μM slices. The filamentous NS1 image (lower panel) shown is a flattened reconstitution from a 10-μM Z stack with 0.6-μM slices in order to highlight the filamentous structures.
Detection of lectin binding.
The DIG glycan differentiation kit (Roche, Indianapolis, IN) was used according to the manufacturer's instructions. Sialic acid linked to the penultimate galactose or N-acetylgalactosamine via an α-2,3 linkage was detected with digoxigenin-conjugated Maackia amurensis agglutinin (MAA) lectin, and α-2,6-linked sialic acid was detected with digoxigenin-conjugated Sambucus nigra agglutinin lectin. Sheep anti-digoxigenin-fluorescein Fab fragments (1:50; Roche) were used to detect lectins for immunofluorescence.
Mouse IFN-β ELISA.
Concentrations of IFN-β were determined by using the mouse IFN-β enzyme-linked immunosorbent assay (ELISA) kit (PBL Biomedical Laboratories) according to the manufacturer's instructions. For mTEC culture experiments, 100 μl of supernatant from the apical and basolateral chambers was assayed by ELISA. Because of the volume differences in the apical versus basolateral chambers, the data are presented as total picograms per sample.
Cytokine bead arrays.
The mouse inflammation cytokine bead array kit (BD Biosciences, San Jose, CA), containing antibodies to IL-6, IL-10, monocyte chemoattractant protein 1 (MCP-1), IFN-γ, TNF-α, and IL-12p70, was used to detect cytokines in the supernatants of mTEC cultures. Fifty microliters of apical or basolateral supernatant from the indicated days were processed according to the manufacturer's instructions by flow cytometry. Because of the volume differences in the apical and basolateral chambers, the data are presented as total picograms per sample.
VSV-GFP L929 bioassay.
L929 murine fibroblasts were pretreated overnight with recombinant mouse IFN-β (PBL Biomedical Laboratories) or basolateral supernatants from mTEC culture infections. Pretreatment inoculum was removed; cells were washed twice with PBS and infected with VSV-GFP at an MOI of 10. Ten hours postinfection, cells were detached with PBS containing 2× trypsin (Sigma, St. Louis, MO), washed twice with PBS, and fixed in PBS containing 1% methanol-free formaldehyde for 15 min at room temperature. Cells were analyzed by flow cytometry (FACSCalibur dual laser flow cytometer; Becton Dickinson) using CellQuest software. Where indicated, 2 μl of antiserum to mouse IFN-α/β (or control sera) was preincubated with basolateral supernatants or recombinant mouse IFN-β for 30 min at room temperature prior to overnight pretreatment.
Infection of BALB/c mice.
Six- to 9-week-old female BALB/c mice were used. Mice were anesthetized with 100 μl of a ketamine (100 mg/kg of body weight)-xylazine (20 mg/kg) cocktail administered via intraperitoneal injection. Mice received a 30-μl intranasal inoculation of virus (virus dosage is indicated in figures) diluted in DMEM with penicillin-streptomycin and 2 μg/ml N-acetyl trypsin (Sigma). Animals were monitored for 21 days postinfection for morbidity and mortality. Mice were weighed daily, and the data were graphed as the percent of weight at the time of infection.
Lung titers of influenza A virus-infected mice.
Whole-lung tissue was harvested from female BALB/c mice infected with 1 × 104 PFU on days 2, 4, and 6 postinfection. Mice were euthanized with 100 μl of a 390-mg/ml solution of sodium pentobarbital via intraperitoneal injection. Tissues were excised and homogenized in a 10% wt/vol solution of PBS (GIBCO, Inc.) and centrifuged twice at 1,200 rpm for 5 min to remove debris. Infectious virus titers were determined by using MDCK cells as previously described (66, 82).
RESULTS
rWSN NS1 R38A is attenuated in growth and plaque size on MDCK cells.
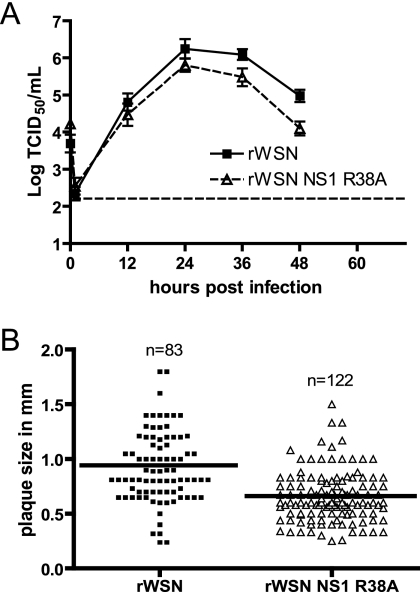
The replication of rWSN NS1 R38A in MDCK cells, a cell line routinely used to grow and plaque influenza A virus, was characterized (Fig. 1A) by performing a multistep growth curve. The rWSN NS1 R38A virus-infected cells produced slightly lower amounts of infectious virus particles than rWSN virus-infected cells did, with titer decreases being more pronounced at 36 and 48 h postinfection (Fig. 1A). Using flow cytometry, the expression levels of NS1 protein at 6 and 9 h postinfection with an MOI of 5 were equivalent in rWSN- and rWSN NS1 R38A-infected cells, indicating that the R38A substitution did not alter NS1 protein expression (data not shown). There was also a small but significant reduction in plaque size for rWSN NS1 R38A compared to that for rWSN (rWSN diameter [± standard deviation {SD}], 0.9346 mm ± 0.03813; rWSN NS1 R38A diameter [± SD], 0.6584 mm ± 0.02110; P < 0.001, two-tailed unpaired t test) (Fig. 1B). These data indicate that values for replication and plaque size in MDCK cells for rWSN NS1 R38A were reduced compared to those for rWSN.
FIG. 1.
rWSN NS1 R38A replication in MDCK cells. (A) Multistep growth curve of rWSN and rWSN NS1 R38A. MDCK cells were infected at an MOI of 0.01, supernatants were collected at the times indicated, and infectious virus titers were determined by 50% tissue culture infective dose (TCID50) assays. The data points are average titers from three separate experiments, and error bars indicate standard errors of the means. The dashed horizontal line indicates the limit of detection. (B) Plaque diameters for rWSN and rWSN NS1 R38A on MDCK cells. The solid line indicates the average plaque size for each group.
rWSN NS1 R38A is attenuated in BALB/c mice.
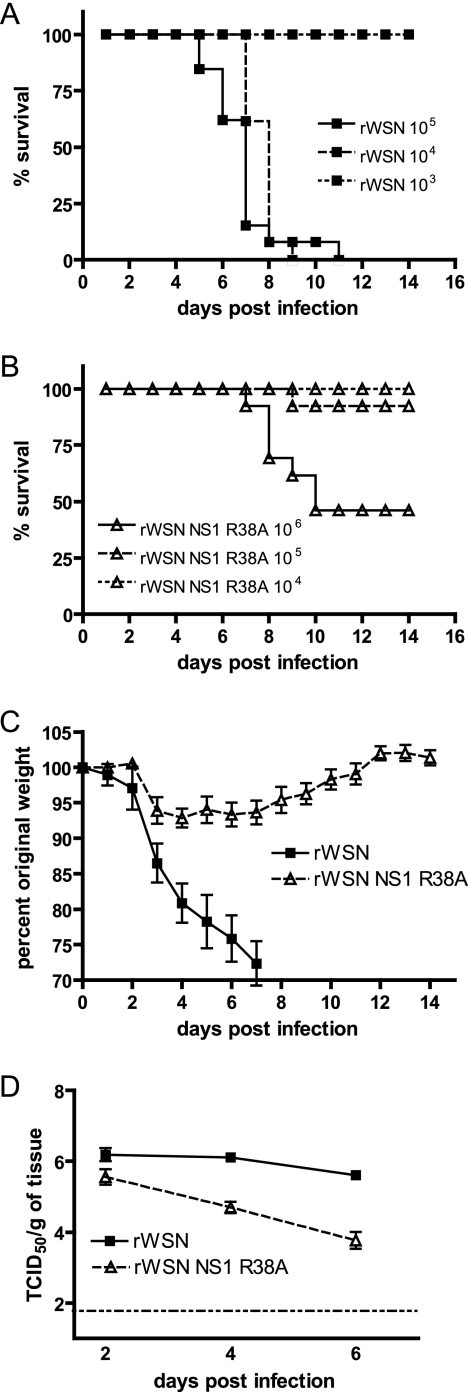
Introducing mutations into the NS1 protein of influenza A virus can lead to dramatic changes in virus virulence (4, 8, 16-18, 21, 22, 24, 30, 32, 33, 36, 37, 41, 52, 54, 59, 60, 72, 73, 75, 79, 81, 87, 91, 97, 99, 100, 103, 103, 107, 108, 113, 114, 118, 124, 125). To determine the effects of the NS1 R38A mutation on influenza A virus virulence, female BALB/c mice were infected by intranasal inoculation with varying doses of rWSN or rWSN NS1 R38A in a volume of 30 μl and were observed for 21 days for lethality and weight loss. The 50% lethal dose for rWSN NS1 R38A (3.0 × 105 PFU) (Fig. 2B) was approximately 2 log units higher than that for rWSN (5.8 × 103 PFU) (Fig. 2A). Transient weight loss was observed in rWSN NS1 R38A-infected mice with original weight levels being reached as early as day 12 postinfection (Fig. 2C). In contrast, rWSN-infected mice displayed marked weight loss and complete mortality by day 9 postinfection at the same virus dose (Fig. 2A and C). The first day of a clear separation in weight loss between the two groups was at day 3 postinfection (mean weight [± SD] for rWSN NS1 R38A, 93.90% ± 1.860%; mean weight [± SD] for rWSN 86.44% ± 0.8570%; P = 0.0013; two-tailed unpaired t test).
FIG. 2.
rWSN NS1 R38A infection of BALB/c mice. Dose-dependent lethality of (A) rWSN and (B) rWSN NS1 R38A in female BALB/c mice. Virus was administered intranasally with the indicated PFU in a volume of 30 μl, and mice were monitored for mortality for 14 days. (C) Weight loss of rWSN- and rWSN NS1 R38A-infected mice after infection with 1 × 104 PFU. Individual mice were weighed daily, and values are expressed as percents of weight at the time of infection. (D) Lung titers of rWSN- and rWSN NS1 R38A-infected mice after infection with 1 × 104 PFU. Error bars indicate standard errors of the means. The dashed horizontal line indicates the limit of detection. Results were reproduced twice.
To specifically address the contribution of the NS1 R38A mutation to virus replication in the respiratory tract, we determined virus lung titers at days 2, 4, and 6 postinfection (Fig. 2D). Compared to wild-type virus, rWSN NS1 R38A viral titers were similar at day 2 postinfection but had declined by several log units on days 4 and 6 postinfection. These data confirm that the NS1 R38A mutation decreases virus mortality and weight loss during infection of BALB/c mice, and rWSN NS1 R38A replication appears to be attenuated in the respiratory tract at later times postinfection. These results are consistent with previous reports demonstrating that mutations in the NS1 RNA binding domain reduce virus virulence in vivo (16).
rWSN NS1 R38A replication and cell tropism in mTEC cultures.
Since rWSN NS1 R38A replication is attenuated in vivo, it is possible that the innate immune responses of the virus-infected epithelial cells could be contributing to the decline in virus replication. In order to evaluate the epithelial-specific innate immune responses generated during influenza A virus infection, we utilized mTEC cultures. We first compared the abilities of rWSN and rWSN NS1 R38A to infect and replicate in mTEC cultures. Differentiated mTEC cultures were infected with a low MOI (3,600 PFU) of rWSN or rWSN NS1 R38A, and infectious virus production in the apical and basolateral supernatants was monitored over a 6-day time course. Influenza A virus budding occurs via the apical surface of polarized airway cells (93), and we have previously reported the detection of virus only in the apical supernatants of mouse and hamster TEC cultures (42, 94). Virus production in the apical supernatant peaked at day 3 postinfection for both viruses and declined thereafter (Fig. 3A). rWSN NS1 R38A-infected cells produced 5-fold-lower to 10-fold-lower amounts of infectious virus particles than did rWSN. Viral antigen (HA) was predominantly expressed in ciliated cells (β-tubulin IV) during infection with either virus (Fig. 3B, upper panels), and there was no significant difference in the number of rWSN NS1 R38A- or rWSN-infected cells (Fig. 3C and D). α2,3-Linked sialic acid, as detected by binding of MAA lectin, is expressed preferentially in mTEC cultures (Fig. 3B, lower panels), and its expression appears to be predominantly in ciliated cells or cells differentiating into a ciliated cell phenotype (42) and is one factor that may limit influenza A virus tropism in mTEC cultures. The rWSN virus preferentially binds to α2,3-linked sialic acid (27, 28, 63), but influenza A virus cell tropism may also be affected by IFN (29, 51) and mutations in the NS1 protein can lead to increased IFN production and signaling (4, 16, 22, 30, 32, 33, 60, 72, 79, 81, 84, 103, 107, 113, 124, 125). Since there are no significant changes in the number of infected cells or in the cell types infected, we conclude that the NS1 R38A mutation does not alter influenza A virus cell tropism but does attenuate virus replication in mTEC cultures.
NS1 subcellular localization in mTECs.
The subcellular localization of NS1 may play an important role in modulating NS1 function and also help explain virus strain-specific activities of the protein (38, 54, 68, 73). The subcellular localization of NS1 and NS1 R38A in mTEC cultures was analyzed by confocal microscopy at day 3 postinfection. WSN NS1 localizes mainly to the cytoplasm of infected cells (Fig. 4, upper panels), and this pattern was also observed with WSN NS1 R38A (Fig. 4, middle panels). These results are in contrast to the NS1 localization patterns observed in MDCK cells, where both NS1 and NS1 R38A localize to both the nucleus and the cytoplasm at 3 h postinfection irrespective of the virus strain used (Fig. 5). Infrequently but consistently, the WSN NS1 R38A protein was detected in filamentous structures in the cytoplasm of infected cells (Fig. 4, lower panels). However, the majority of the rWSN NS1 R38A-infected cells displayed a NS1 localization pattern indistinguishable from that observed in rWSN-infected cells at this particular time postinfection, suggesting that the NS1 R38A mutation does not significantly alter the subcellular localization of the protein under the assayed conditions in infected mTEC cultures. This is in contrast to the NS1 localization patterns observed in MDCK cells, where both NS1 and NS1 R38A localize to both the nucleus and the cytoplasm at 3 h postinfection irrespective of the virus strain used (Fig. 5).
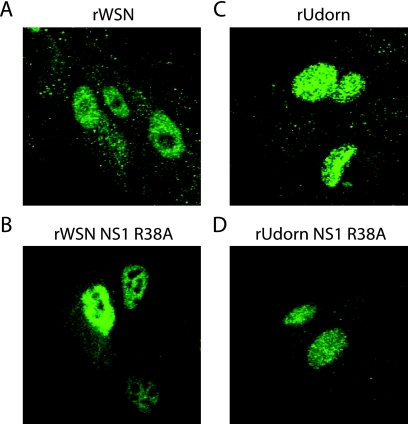
FIG. 5.
NS1 subcellular localization in MDCK cells. MDCK cells were infected at an MOI of 5 with (A) rWSN, (B) rWSN NS1 R38A, (C) rUdorn, or (D) rUdorn NS1 R38A, fixed at 3 h postinfection, immunostained with anti-NS1 antibody, and analyzed by confocal microscopy. All images were taken at ×63 magnification. The R38A mutation was introduced into the NS segment of rUdorn according to established protocols (112).
Increased cytokine production during rWSN NS1 R38A infection.
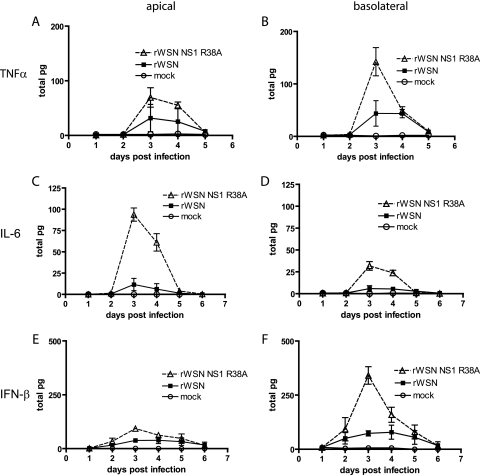
The NS1 protein is the major viral protein responsible for inhibiting the host cell response to influenza A virus infection (4, 8, 16, 18, 22, 24, 32, 33, 36, 53, 56, 59, 60, 72, 73, 79, 81, 89, 99, 100, 103, 108, 113, 114, 125). To assess the effect of the NS1 R38A mutation on virus-induced cytokine production, apical and basolateral supernatants from rWSN- and rWSN NS1 R38A-infected mTEC cultures were monitored for the secretion of inflammatory cytokines and IFN-β. The cytokines IL-10, MCP-1, IFN-γ, and IL-12p70 were not induced by infection with either virus (data not shown). Infection with either virus resulted in increased production of TNF-α (Fig. 6A and B), IL-6 (Fig. 6C and D), and IFN-β (Fig. 6E and F), but significantly higher cytokine levels were found in rWSN NS1 R38A-infected cultures. The peak in production for all three cytokines occurred at day 3 postinfection, correlating with the peak viral titer (Fig. 3). Interestingly, the cytokines appeared to be secreted in a polarized manner, with IFN-β and TNF-α being predominantly detected in the basolateral supernatants (Fig. 6B and F), while IL-6 was secreted primarily into the apical supernatant (Fig. 6C). Taken together, the data indicate that infection with rWSN NS1 R38A leads to dramatic changes in the expression of certain proinflammatory and antiviral cytokines.
FIG. 6.
Cytokine production in virus-infected mTEC cultures. Apical and basolateral supernatants were analyzed for the presence of TNF-α, IFN-γ, IL-6, IL-10, MCP-1, and IL-12p70 by cytokine bead array and for the presence of IFN-β by ELISA. Shown are results for TNF-α (A and B), IL-6 (C and D), and IFN-β (E and F) cytokine production in the apical (A, C, and E) and basolateral (B, D, and F) chambers. All other cytokines tested were at levels indistinguishable from those of mock-infected cultures (data not shown). The levels of cytokines present in the apical (left column) and basolateral (right column) chambers were normalized to total picograms to account for the differences in apical (100 μl) and basolateral (500 μl) chamber volumes. Data points are the averages from one experiment, and standard errors of the means (error bars) are shown. Results were replicated at least four times.
Antiviral activity of influenza A virus-induced cytokines.
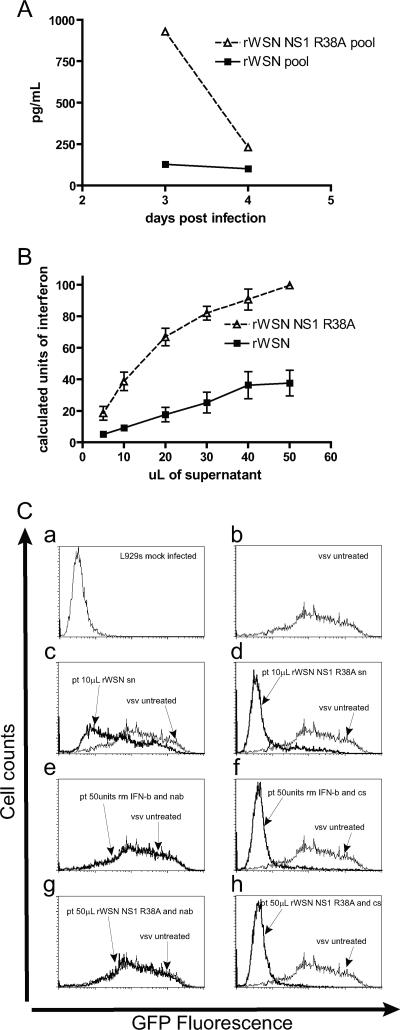
Cytokines such as TNF-α and IFN-β are known to possess antiviral activities (25, 26, 44, 45, 62, 70, 71, 101, 119, 128). In order to determine whether the cytokines secreted in response to influenza virus infection had antiviral activities, we examined the effects of pretreatment with cytokine-rich, basolateral supernatants from influenza virus-infected mTEC cultures on the replication of VSV-GFP. VSV replication is known to be sensitive to the presence of type I interferon (23, 34, 109) and, thus, is a useful tool for determining the amount of IFN activity present in virus-infected supernatants. Mouse fibroblast L929 cells were pretreated overnight with recombinant murine IFN-β or with samples of basolateral supernatants from rWSN- or rWSN NS1 R38A-infected mTEC cultures and were infected with a high MOI of VSV expressing GFP (5), and GFP expression was quantified by flow cytometry at 10 h postinfection. The inhibition of VSV gene expression was observed with recombinant IFN-β, and a dose response curve was determined (data not shown). By comparing the amount of VSV inhibition observed with the standardized amounts of IFN-β to the inhibition observed after pretreatment with varying volumes of pooled mTEC cytokine-rich supernatants from day 3, a relative measure of IFN-β activity present in rWSN- or rWSN NS1 R38A-infected cell supernatants was determined (Fig. 7B). For example, 10 μl of supernatant from rWSN-infected mTEC cultures had modest effects on the replication of VSV-GFP (Fig. 7C, panel c), while the same amount of supernatant from rWSN NS1 R38A-infected cultures resulted in a dramatic reduction of VSV-GFP replication (Fig. 7C, panel d). IFN-β activity increased 3.5- to 5-fold overall for day 3 basolateral supernatants from rWSN NS1 R38A-infected cells relative to those from rWSN-infected cells. IFN-β appeared to be the cytokine primarily responsible for this effect, as the addition of type I IFN neutralizing serum restored VSV-GFP gene expression (Fig. 7C, panels e and g) compared to the case for the control serum (Fig. 7C, panels f and h). Taken together, these data indicate that IFN-β production during rWSN NS1 R38A infection is increased in amount (Fig. 7A) and biological activity (Fig. 7B and C) and that the additional cytokines detected (TNF-α and IL-6) appear to have a minimal effect on VSV-GFP replication in L929 cells.
FIG. 7.
Antiviral activity of basolateral supernatants from rWSN- and rWSN NS1 R38A-infected mTEC cultures. (A) IFN-β ELISA of pooled basolateral samples from rWSN- and rWSN NS1 R38A-infected cultures at days 3 and 4 postinfection. (B) Calculated units of type I IFN activity in rWSN- and rWSN NS1 R38A-infected mTEC culture basolateral supernatants from day 3 postinfection. L929 murine fibroblasts were pretreated overnight with the indicated volumes of basolateral supernatant, infected with VSV-GFP at an MOI of 10, and harvested 10 h postinfection for analysis by flow cytometry. The amount of VSV-GFP inhibition observed with basolateral supernatants was compared to that with a mouse recombinant IFN-β standard curve (not shown) to calculate approximate units of type I interferon. (C) Flow cytometry for VSV-GFP gene expression. (Panel a) Untreated L929 cells. (Panel b) VSV-GFP gene expression on L929 cells. (Panels c and d) Pretreatment with 10 μl of basolateral supernatant from (c) rWSN- or (d) rWSN NS1 R38A-infected mTEC cultures. (Panel e) Pretreatment with 50 U of mouse recombinant IFN-β and type I interferon neutralizing sera. (Panel f) Pretreatment with 50 U of recombinant mouse IFN-β and control sera. (Panel g) Pretreatment with 50 μl of rWSN NS1 R38A basolateral supernatants and type I interferon neutralizing sera. (Panel h) Pretreatment with 50 μl of rWSN NS1 R38A basolateral supernatants and control sera. These experiments were replicated in triplicate using two different pools of basolateral supernatant. pt, pretreatment; sn, supernatant; nab, type I interferon neutralizing antibody; cs, control serum.
rWSN NS1 R38A is sensitive to cytokine pretreatment.
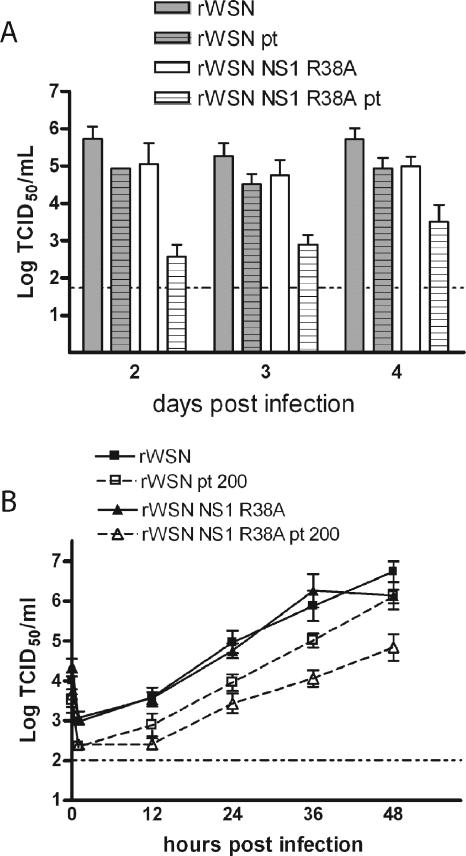
Cytokines such as IFN-β have clearly defined antiviral activities, while cytokines such as TNF-α and IL-6 may alter virus replication directly (101, 119) but also serve to recruit or activate immune cells. To determine the effects of cytokine pretreatment on influenza A virus replication, naïve mTEC cultures were pretreated overnight with cytokine-rich supernatants from rWSN NS1 R38A-infected cells, infected with a low MOI of rWSN or rWSN NS1 R38A, and monitored for infectious virus production. Pretreatment had minimal effects on rWSN replication (Fig. 8A), but rWSN NS1 R38A titers were drastically reduced at all times sampled, indicating that the cytokine-rich supernatant was capable of attenuating only rWSN NS1 R38A infection (Fig. 8A).
FIG. 8.
Influenza A virus sensitivity to cytokine pretreatment. (A) Cytokine pretreatment in mTEC cultures. Naïve mTEC cultures were pretreated overnight with 500 μl of the day 3 basolateral supernatants from rWSN NS1 R38A infection. Cells were then infected with rWSN or rWSN NS1 R38A at an MOI of 0.01 (3,600 PFU), and infectious virus production was monitored for 4 days. Infectious virus titers were determined by 50% tissue culture infective dose (TCID50) assay. Shown are the apical titers for each virus. Virus was not detected in the basolateral supernatants during infection or in the supernatants used for pretreatment. Data points are the averages of three replicates, shown with standard errors of the means. Pretreatment had minimal effects on rWSN replication (gray bars versus gray hatched bars), but rWSN NS1 R38A titers were drastically reduced at all times sampled, indicating that the cytokine-rich supernatant was capable of attenuating only rWSN NS1 R38A infection (white bars versus white hatched bars). (B) Vero cells were pretreated overnight with 200 U of recombinant human IFN-β and then infected with rWSN or rWSN NS1 R38A at an MOI of 0.01. Infectious virus titers were determined by TCID50 assay at the times indicated, and standard errors of the means (error bars) are shown. The data was replicated twice.
Since IFN-β has known antiviral activity, we wanted to determine whether this cytokine could be responsible for the inhibition of rWSN NS1 R38A infection. Vero cells lack the type I interferon α and β genes (15, 19, 74) but can respond to exogenous interferon. Therefore, the initial effects of interferon pretreatment versus the subsequent effects of type I interferon autocrine and paracrine signaling can be separated. Unlike other cell types (Fig. 1A and 3A), the viruses replicated to equivalent titers on Vero cells (Fig. 8B), suggesting that the endogenous production of type I interferon may be important in limiting rWSN NS1 R38A replication. Overnight treatment of Vero cells with human recombinant IFN-β resulted in decreased replication with both viruses; however, the rWSN NS1 R38A virus titers were reduced to a larger extent than those of rWSN (Fig. 8B). The magnitude of virus replication inhibition was not as great as that observed in the mTEC cultures, suggesting either that there is a role for other cytokines and/or endogenous IFN-β production in inhibiting rWSN NS1 R38A replication or that there are cell type-specific effects of IFN-β on influenza A virus replication. The pretreatment experiments on both mTEC cultures and Vero cells demonstrate that rWSN NS1 R38A is more sensitive to the effects of cytokines such as IFN-β and that the autocrine and paracrine IFN-β signaling pathways may be important for enhancing these effects in mTEC cultures.
DISCUSSION
Primary airway epithelial cell models have been used to identify important features of virus replication for a number of respiratory viruses (3, 7, 11, 20, 39, 42, 49, 64, 65, 69, 78, 85, 86, 95, 104-106, 110, 117, 121, 129, 131, 132, 134, 135), and here we present the first study using primary airway epithelial cell cultures to analyze influenza A virus NS1 function in the regulation of airway innate immune responses. Increased production of TNF-α, IL-6, and IFN-β during rWSN NS1 R38A infection of mTEC cultures suggests that NS1 is a critical regulator of cytokine induction in primary airway cells and likely regulates multiple pathways to achieve its inhibitory effects.
The NS1 RNA binding domain is important for the inhibition of transcription factors IRF-3 (113), NF-κB (125), and ATF2/c-Jun (60), components that are necessary (along with IRF-7) to fully activate the IFN-β promoter (47, 55, 61, 96, 98, 115, 116, 126). The activation of NF-κB also leads to transcriptional upregulation of proinflammatory cytokines, including TNF-α and IL-6 (40, 50). The activation of NF-κB and IRF-3 can occur through multiple signaling pathways, including Toll-like receptor 3, RIG-I, melanoma differentiation-associated gene MDA-5, and PKR (NF-κB only) (40, 102). NS1 can interact with RIG-I and inhibit the RIG-I/IPS-1:CARD/IRF-3 signaling pathway (72), along with inhibiting the activation of PKR (52, 114). Studies of cell lines or mouse strains that are deficient in these signaling pathways have shown that all are important sensors of virus infection, though some of the pathways are particularly sensitive to a select group of viruses. Whether there are any cell type-specific roles for the cellular RNA sensors is not yet clear, but mTEC cultures should provide a useful platform for determining the role of these pathways in the antiviral response of respiratory epithelium to influenza virus.
The NS1 protein is localized primarily in the cytoplasm of virus-infected mTEC cultures. Previous reports examining NS1 localization in MDCK cells have found that the protein is both in the cytoplasm and in the nucleus during infection (31, 68, 73). We analyzed NS1 localization during infection of MDCK cells using rWSN and rUdorn and found that NS1 subcellular localization is dependent on several factors, including the virus strain used, the amount of NS1 present, the cell type utilized and the time postinfection (Fig. 5 and data not shown). The most conserved NLS of the NS1 protein appears to coincide with the RNA binding domain (31, 123). Mutating basic amino acid residues in the NLS/RNA binding region have been reported to affect nuclear versus cytoplasmic localization differently in rWSN versus rUdorn, owing to a second NLS (NLS2) sequence near the carboxy terminal of the NS1 encoded by rUdorn (31, 68, 73). While mutations in the NLS of WSN NS1 lead to a greater amount of cytoplasmic NS1, the protein is not entirely excluded from the nucleus (68, 73), suggesting that there are other yet undefined sequences in NS1 that play a role in nuclear localization. In addition, because NLS2 is not present in a significant number of pathogenic influenza A virus strains (68), it is difficult to determine how critical this sequence is to viral pathogenesis. It is also possible, if not likely, that the subcellular localization of NS1 will be cell type dependent. Factors such as the state of cell polarity as well as the expression of cell type-dependent proteins may significantly alter NS1 subcellular localization. Since changes in intracellular localization as well as in the double-stranded RNA binding activity of NS1 can be attributed to the same protein sequences, discerning the individual contributions of both activities to a specific phenotype will be a difficult but important task.
Influenza A virus infection of mTEC cultures may lead to the production of other as-yet-unknown cytokines. While three important antiviral/proinflammatory components have been identified, we have taken a targeted approach to cytokine screening in this study. Experiments are underway to provide a more comprehensive view of the cytokines and antiviral molecules produced in mTEC cultures infected with both wild-type and genetically engineered influenza A viruses. An additional likely candidate is IL-8, which is secreted during influenza A virus infection of primary human airway cells (3, 11). However, whether NS1 protein is important for the regulation of IL-8 (or the murine equivalents KC and MIP-2) in airway cells, as it seems to be for IFN-β, TNF-α, and IL-6, is not known. IL-8 induction occurs via an NF-κB-dependent pathway (92), and it is therefore possible that NS1 may globally regulate TNF-α, IL-6, and IL-8 production by inhibiting NF-κB signaling pathways.
IFN-β, TNF-α, and IL-6 are released in a directional manner during rWSN NS1 R38A mTEC culture infection. Polarized secretion of cytokines during virus infection has been reported for similar primary differentiated airway systems (69), and this again highlights an advantage of primary airway systems over cell lines. Transformed cell lines used for virus-induced cytokine studies often lack the unique properties of polarization and cell type heterogeneity that the primary differentiated cultures provide. In addition, polarized cytokine secretion in primary cultures provides a framework to consider how immune cells might be directionally recruited to the site of infection in vivo (39), i.e., from the lumen of the lung versus the basement membrane and underlying vascular system. The specific cell type(s) responsible for inflammatory cytokine production during rWSN NS1 R38A infection of mTEC cultures is not known at this time. Ciliated cells were identified as the inflammatory cytokine secretors during RSV infection (69), and RSV exclusively infects ciliated cells in that system. Thus, it is possible, if not likely, that the influenza virus-infected ciliated cells are secreting the antiviral/inflammatory cytokines.
Our studies suggest that, in addition to a reduced ability to inhibit cytokine production, rWSN NS1 R38A is more sensitive to cytokine pretreatment than wild-type virus is in both the mTEC cultures and the Vero cell line. This result is in agreement with those of similar studies that found that the NS1 R38A mutation in the rUdorn background resulted in a virus more sensitive to IFN-β pretreatment on A549 cells, a type II pneumocyte-derived cell line (73). Thus, it appears that the cytokine sensitivity resulting from the R38A mutation is not strain or cell line specific and is perhaps a general property of all NS1 proteins. Whether the mechanism(s) responsible for increased cytokine production in rWSN NS1 R38A-infected cultures is also involved in the increased sensitivity of this virus to cytokine pretreatment is not clear. It is clear, however, that type I interferon pretreatment is inhibitory to viruses possessing the NS1 R38A mutation (this study; 73), and further studies will be needed to determine the effects of NS1 function during pretreatment versus naïve infection. The use of differentiated, primary cell cultures, such as murine, hamster, and human TECs in studies of influenza A virus replication as well as host response to infection should provide a wealth of relevant information that will increase our understanding of virus-epithelial cell interactions.
Acknowledgments
We acknowledge the Molecular Microbiology Imaging Facility, Washington University School of Medicine, for technical support and all the members of the Pekosz laboratory for insightful discussions and comments.
This work was supported by the Department of Health and Human Services, Public Health Services grant AI053629 (A.P.).
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Alonso-Caplen, F. V., M. E. Nemeroff, Y. Qiu, and R. M. Krug. 1992. Nucleocytoplasmic transport: the influenza virus NS1 protein regulates the transport of spliced NS2 mRNA and its precursor NS1 mRNA. Genes Dev. 6:255-267. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2004. Update: influenza-associated deaths reported among children aged <18 years—United States, 2003-04 influenza season. Ann. Pharmacother. 38:367-368. [DOI] [PubMed] [Google Scholar]
- 3.Arndt, U., G. Wennemuth, P. Barth, M. Nain, Y. Al-Abed, A. Meinhardt, D. Gemsa, and M. Bacher. 2002. Release of macrophage migration inhibitory factor and CXCL8/interleukin-8 from lung epithelial cells rendered necrotic by influenza A virus infection. J. Virol. 76:9298-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann, M., A. Garcia-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boritz, E., J. Gerlach, J. E. Johnson, and J. K. Rose. 1999. Replication-competent rhabdoviruses with human immunodeficiency virus type 1 coats and green fluorescent protein: entry by a pH-independent pathway. J. Virol. 73:6937-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, L. E., V. S. Hinshaw, and R. G. Webster. 1983. Antigenic variation in the influenza A virus nonstructural protein, NS1. Virology 130:134-143. [DOI] [PubMed] [Google Scholar]
- 7.Chan, M. C., C. Y. Cheung, W. H. Chui, S. W. Tsao, J. M. Nicholls, Y. O. Chan, R. W. Chan, H. T. Long, L. L. Poon, Y. Guan, and J. S. Peiris. 2005. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir. Res. 6:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, C. Y., L. L. Poon, A. S. Lau, W. Luk, Y. L. Lau, K. F. Shortridge, S. Gordon, Y. Guan, and J. S. Peiris. 2002. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet 360:1831-1837. [DOI] [PubMed] [Google Scholar]
- 10.Chien, C. Y., R. Tejero, Y. Huang, D. E. Zimmerman, C. B. Rios, R. M. Krug, and G. T. Montelione. 1997. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat. Struct. Biol. 4:891-895. [DOI] [PubMed] [Google Scholar]
- 11.Choi, A. M., and D. B. Jacoby. 1992. Influenza virus A infection induces interleukin-8 gene expression in human airway epithelial cells. FEBS Lett. 309:327-329. [DOI] [PubMed] [Google Scholar]
- 12.Cox, N. J., and K. Subbarao. 2000. Global epidemiology of influenza: past and present. Annu. Rev. Med. 51:407-421. [DOI] [PubMed] [Google Scholar]
- 13.de Jong, M. D., C. P. Simmons, T. T. Thanh, V. M. Hien, G. J. Smith, T. N. Chau, D. M. Hoang, N. V. Chau, T. H. Khanh, V. C. Dong, P. T. Qui, B. Van Cam, Q. D. Ha, Y. Guan, J. S. Peiris, N. T. Chinh, T. T. Hien, and J. Farrar. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Martin, S., and A. Nicoll. 2005. H5N1 avian influenza: update on the global situation. Eurosurveillance 10:E051215.1. http://www.eurosurveillance.org/ew/2005/051215.asp#1. [DOI] [PubMed] [Google Scholar]
- 15.Diaz, M. O., S. Ziemin, M. M. Le Beau, P. Pitha, S. D. Smith, R. R. Chilcote, and J. D. Rowley. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. 85:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donelan, N. R., C. F. Basler, and A. Garcia-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77:13257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egorov, A., S. Brandt, S. Sereinig, J. Romanova, B. Ferko, D. Katinger, A. Grassauer, G. Alexandrova, H. Katinger, and T. Muster. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehrhardt, C., T. Wolff, S. Pleschka, O. Planz, W. Beermann, J. G. Bode, M. Schmolke, and S. Ludwig. 2007. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81:3058-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43:247-252. [DOI] [PubMed] [Google Scholar]
- 20.Endo, Y., K. N. Carroll, M. R. Ikizler, and P. F. Wright. 1996. Growth of influenza A virus in primary, differentiated epithelial cells derived from adenoids. J. Virol. 70:2055-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falcón, A. M., A. Fernandez-Sesma, Y. Nakaya, T. M. Moran, J. Ortin, and A. Garcia-Sastre. 2005. Attenuation and immunogenicity in mice of temperature-sensitive influenza viruses expressing truncated NS1 proteins. J. Gen. Virol. 86:2817-2821. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez-Sesma, A., S. Marukian, B. J. Ebersole, D. Kaminski, M.-S. Park, T. Yuen, S. C. Sealfon, A. Garcia-Sastre, and T. M. Moran. 2006. Influenza virus evades innate and adaptive immunity via the NS1 protein. J. Virol. 80:6295-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira, P. C., M. L. Peixoto, M. A. Silva, and R. R. Golgher. 1979. Assay of human interferon in Vero cells by several methods. J. Clin. Microbiol. 9:471-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fortes, P., A. Beloso, and J. Ortin. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 13:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman, R. M. 1977. Antiviral activity of interferons. Bacteriol. Rev. 41:543-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman, R. M. 1967. Interferon binding: the first step in establishment of antiviral activity. Science 156:1760-1761. [DOI] [PubMed] [Google Scholar]
- 27.Gambaryan, A. S., J. S. Robertson, and M. N. Matrosovich. 1999. Effects of egg-adaptation on the receptor-binding properties of human influenza A and B viruses. Virology 258:232-239. [DOI] [PubMed] [Google Scholar]
- 28.Gambaryan, A. S., A. B. Tuzikov, V. E. Piskarev, S. S. Yamnikova, D. K. Lvov, J. S. Robertson, N. V. Bovin, and M. N. Matrosovich. 1997. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-acetyllactosamine). Virology 232:345-350. [DOI] [PubMed] [Google Scholar]
- 29.García-Sastre, A., R. K. Durbin, H. Zheng, P. Palese, R. Gertner, D. E. Levy, and J. E. Durbin. 1998. The role of interferon in influenza virus tissue tropism. J. Virol. 72:8550-8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 31.Greenspan, D., P. Palese, and M. Krystal. 1988. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J. Virol. 62:3020-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo, Z., L. M. Chen, H. Zeng, J. A. Gomez, J. Plowden, T. Fujita, J. M. Katz, R. O. Donis, and S. Sambhara. 2007. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 36:263-269. [DOI] [PubMed] [Google Scholar]
- 33.Hale, B. G., D. Jackson, Y. H. Chen, R. A. Lamb, and R. E. Randall. 2006. Influenza A virus NS1 protein binds p85β and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. USA 103:14194-14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hallum, J. V., H. R. Thacore, and J. S. Youngner. 1970. Factors affecting the sensitivity of different viruses to interferon. J. Virol. 6:156-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatada, E., and R. Fukuda. 1992. Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol. 73:3325-3329. [DOI] [PubMed] [Google Scholar]
- 36.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayman, A., S. Comely, A. Lackenby, L. C. S. Hartgroves, S. Goodbourn, J. W. McCauley, and W. S. Barclay. 2007. NS1 proteins of avian influenza A viruses can act as antagonists of the human alpha/beta interferon response. J. Virol. 81:2318-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayman, A., S. Comely, A. Lackenby, S. Murphy, J. McCauley, S. Goodbourn, and W. Barclay. 2006. Variation in the ability of human influenza A viruses to induce and inhibit the IFN-β pathway. Virology 347:52-64. [DOI] [PubMed] [Google Scholar]
- 39.Herold, S., W. von Wulffen, M. Steinmueller, S. Pleschka, W. A. Kuziel, M. Mack, M. Srivastava, W. Seeger, U. A. Maus, and J. Lohmeyer. 2006. Alveolar epithelial cells direct monocyte transepithelial migration upon influenza virus infection: impact of chemokines and adhesion molecules. J. Immunol. 177:1817-1824. [DOI] [PubMed] [Google Scholar]
- 40.Hiscott, J., T. L. Nguyen, M. Arguello, P. Nakhaei, and S. Paz. 2006. Manipulation of the nuclear factor-κB pathway and the innate immune response by viruses. Oncogene 25:6844-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hyland, L., R. Webby, M. R. Sandbulte, B. Clarke, and S. Hou. 2006. Influenza virus NS1 protein protects against lymphohematopoietic pathogenesis in an in vivo mouse model. Virology 349:156-163. [DOI] [PubMed] [Google Scholar]
- 42.Ibricevic, A., A. Pekosz, M. J. Walter, C. Newby, J. T. Battaile, E. G. Brown, M. J. Holtzman, and S. L. Brody. 2006. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 80:7469-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Influenza Team of the European Centre for Disease Prevention and Control. 2006. Highly pathogenic avian influenza A/H5N1—update and overview of 2006. Eurosurveillance 11:E061221.1. http://www.eurosurveillance.org/ew/2006/061221.asp#1. [DOI] [PubMed] [Google Scholar]
- 44.Ito, M., T. Nakano, T. Kamiya, K. Kitamura, T. Ihara, H. Kamiya, and M. Sakurai. 1991. Effects of tumor necrosis factor alpha on replication of varicella-zoster virus. Antivir. Res. 15:183-192. [DOI] [PubMed] [Google Scholar]
- 45.Ito, M., and J. A. O'Malley. 1987. Antiviral effects of recombinant human tumor necrosis factor. Lymphokine Res. 6:309-318. [PubMed] [Google Scholar]
- 46.Kaufmann, A., R. Salentin, R. G. Meyer, D. Bussfeld, C. Pauligk, H. Fesq, P. Hofmann, M. Nain, D. Gemsa, and H. Sprenger. 2001. Defense against influenza A virus infection: essential role of the chemokine system. Immunobiology 204:603-613. [DOI] [PubMed] [Google Scholar]
- 47.Kim, T. K., and T. Maniatis. 1997. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol. Cell 1:119-129. [DOI] [PubMed] [Google Scholar]
- 48.Kochs, G., A. Garcia-Sastre, and L. Martinez-Sobrido. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 81:7011-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kogure, T., T. Suzuki, T. Takahashi, D. Miyamoto, K. Hidari, C.-T. Guo, T. Ito, Y. Kawaoka, and Y. Suzuki. 2006. Human trachea primary epithelial cells express both sialyl(α2-3)Gal receptor for human parainfluenza virus type 1 and avian influenza viruses, and sialyl(α2-6)Gal receptor for human influenza viruses. Glycoconj. J. 23:101-106. [DOI] [PubMed] [Google Scholar]
- 50.Kulms, D., and T. Schwarz. 2006. NF-κB and cytokines. Vitam. Horm. 74:283-300. [DOI] [PubMed] [Google Scholar]
- 51.Lenschow, D. J., C. Lai, N. Frias-Staheli, N. V. Giannakopoulos, A. Lutz, T. Wolff, A. Osiak, B. Levine, R. E. Schmidt, A. Garcia-Sastre, D. A. Leib, A. Pekosz, K. P. Knobeloch, I. Horak, and H. W. Virgin IV. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. USA 104:1371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, S., J.-Y. Min, R. M. Krug, and G. C. Sen. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13-21. [DOI] [PubMed] [Google Scholar]
- 53.Li, Y., Y. Yamakita, and R. M. Krug. 1998. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc. Natl. Acad. Sci. USA 95:4864-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li, Z., Y. Jiang, P. Jiao, A. Wang, F. Zhao, G. Tian, X. Wang, K. Yu, Z. Bu, and H. Chen. 2006. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J. Virol. 80:11115-11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin, R., P. Genin, Y. Mamane, and J. Hiscott. 2000. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 20:6342-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lipatov, A. S., S. Andreansky, R. J. Webby, D. J. Hulse, J. E. Rehg, S. Krauss, D. R. Perez, P. C. Doherty, R. G. Webster, and M. Y. Sangster. 2005. Pathogenesis of Hong Kong H5N1 influenza virus NS gene reassortants in mice: the role of cytokines and B- and T-cell responses. J. Gen. Virol. 86:1121-1130. [DOI] [PubMed] [Google Scholar]
- 57.Liu, J., P. A. Lynch, C. Y. Chien, G. T. Montelione, R. M. Krug, and H. M. Berman. 1997. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat. Struct. Biol. 4:896-899. [DOI] [PubMed] [Google Scholar]
- 58.Lu, X., T. M. Tumpey, T. Morken, S. R. Zaki, N. J. Cox, and J. M. Katz. 1999. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 73:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu, Y., X. Y. Qian, and R. M. Krug. 1994. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 8:1817-1828. [DOI] [PubMed] [Google Scholar]
- 60.Ludwig, S., X. Wang, C. Ehrhardt, H. Zheng, N. Donelan, O. Planz, S. Pleschka, A. Garcia-Sastre, G. Heins, and T. Wolff. 2002. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 76:11166-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 62.Matikainen, S., J. Siren, J. Tissari, V. Veckman, J. Pirhonen, M. Severa, Q. Sun, R. Lin, S. Meri, G. Uze, J. Hiscott, and I. Julkunen. 2006. Tumor necrosis factor alpha enhances influenza A virus-induced expression of antiviral cytokines by activating RIG-I gene expression. J. Virol. 80:3515-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matrosovich, M., A. Tuzikov, N. Bovin, A. Gambaryan, A. Klimov, M. R. Castrucci, I. Donatelli, and Y. Kawaoka. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 74:8502-8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matrosovich, M. N., T. Y. Matrosovich, T. Gray, N. A. Roberts, and H.-D. Klenk. 2004. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 78:12665-12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matrosovich, M. N., T. Y. Matrosovich, T. Gray, N. A. Roberts, and H. D. Klenk. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 101:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCown, M. F., and A. Pekosz. 2006. Distinct domains of the influenza A virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 80:8178-8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCown, M. F., and A. Pekosz. 2005. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 79:3595-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melén, K., L. Kinnunen, R. Fagerlund, N. Ikonen, K. Y. Twu, R. M. Krug, and I. Julkunen. 2007. Nuclear and nucleolar targeting of influenza A virus NS1A protein: striking differences between different virus subtypes. J. Virol. 81:5995-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mellow, T. E., P. C. Murphy, J. L. Carson, T. L. Noah, L. Zhang, and R. J. Pickles. 2004. The effect of respiratory synctial virus on chemokine release by differentiated airway epithelium. Exp. Lung Res. 30:43-57. [DOI] [PubMed] [Google Scholar]
- 70.Mestan, J., M. Brockhaus, H. Kirchner, and H. Jacobsen. 1988. Antiviral activity of tumour necrosis factor. Synergism with interferons and induction of oligo-2′,5′-adenylate synthetase. J. Gen. Virol. 69:3113-3120. [DOI] [PubMed] [Google Scholar]
- 71.Mestan, J., W. Digel, S. Mittnacht, H. Hillen, D. Blohm, A. Moller, H. Jacobsen, and H. Kirchner. 1986. Antiviral effects of recombinant tumour necrosis factor in vitro. Nature 323:816-819. [DOI] [PubMed] [Google Scholar]
- 72.Mibayashi, M., L. Martinez-Sobrido, Y.-M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Min, J.-Y., and R. M. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA 103:7100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mosca, J. D., and P. M. Pitha. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 76.Nemeroff, M. E., X. Y. Qian, and R. M. Krug. 1995. The influenza virus NS1 protein forms multimers in vitro and in vivo. Virology 212:422-428. [DOI] [PubMed] [Google Scholar]
- 77.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Newby, C. M., R. K. Rowe, and A. Pekosz. 2006. Influenza A virus infection of primary differentiated airway epithelial cell cultures derived from Syrian golden hamsters. Virology 354:80-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307:386-395. [DOI] [PubMed] [Google Scholar]
- 80.Novak, M., Z. Moldoveanu, D. P. Schafer, J. Mestecky, and R. W. Compans. 1993. Murine model for evaluation of protective immunity to influenza virus. Vaccine 11:55-60. [DOI] [PubMed] [Google Scholar]
- 81.Opitz, B., A. Rejaibi, B. Dauber, J. Eckhard, M. Vinzing, B. Schmeck, S. Hippenstiel, N. Suttorp, and T. Wolff. 2007. IFN-β induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell. Microbiol. 9:930-938. [DOI] [PubMed] [Google Scholar]
- 82.Paterson, R. G., and R. A. Lamb. 1993. The molecular biology of influenza viruses and paramyxoviruses, p. 35-73, Molecular virology: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 83.Peiris, J. S., W. C. Yu, C. W. Leung, C. Y. Cheung, W. F. Ng, J. M. Nicholls, T. K. Ng, K. H. Chan, S. T. Lai, W. L. Lim, K. Y. Yuen, and Y. Guan. 2004. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 363:617-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 85.Pickles, R. J., J. A. Fahrner, J. M. Petrella, R. C. Boucher, and J. M. Bergelson. 2000. Retargeting the coxsackievirus and adenovirus receptor to the apical surface of polarized epithelial cells reveals the glycocalyx as a barrier to adenovirus-mediated gene transfer. J. Virol. 74:6050-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pickles, R. J., D. McCarty, H. Matsui, P. J. Hart, S. H. Randell, and R. C. Boucher. 1998. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J. Virol. 72:6014-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qian, X., F. Alonso-Caplen, and R. Krug. 1994. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 68:2433-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qian, X. Y., C. Y. Chien, Y. Lu, G. T. Montelione, and R. M. Krug. 1995. An amino-terminal polypeptide fragment of the influenza virus NS1 protein possesses specific RNA-binding activity and largely helical backbone structure. RNA 1:948-956. [PMC free article] [PubMed] [Google Scholar]
- 89.Qiu, Y., and R. Krug. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J. Virol. 68:2425-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qiu, Y., M. Nemeroff, and R. M. Krug. 1995. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA 1:304-316. [PMC free article] [PubMed] [Google Scholar]
- 91.Quinlivan, M., D. Zamarin, A. Garcia-Sastre, A. Cullinane, T. Chambers, and P. Palese. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 79:8431-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roebuck, K. A. 1999. Regulation of interleukin-8 gene expression. J. Interferon Cytokine Res. 19:429-438. [DOI] [PubMed] [Google Scholar]
- 93.Roth, M. G., J. P. Fitzpatrick, and R. W. Compans. 1979. Polarity of influenza and vesicular stomatitis virus maturation in MDCK cells: lack of a requirement for glycosylation of viral glycoproteins. Proc. Natl. Acad. Sci. USA 76:6430-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rowe, R. K., S. L. Brody, and A. Pekosz. 2004. Differentiated cultures of primary hamster tracheal airway epithelial cells. In Vitro Cell. Dev. Biol. Anim. 40:303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rowe, R. K., and A. Pekosz. 2006. Bidirectional virus secretion and nonciliated cell tropism following Andes virus infection of primary airway epithelial cell cultures. J. Virol. 80:1087-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sato, M., N. Tanaka, N. Hata, E. Oda, and T. Taniguchi. 1998. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett. 425:112-116. [DOI] [PubMed] [Google Scholar]
- 97.Satterly, N., P. L. Tsai, J. van Deursen, D. R. Nussenzveig, Y. Wang, P. A. Faria, A. Levay, D. E. Levy, and B. M. Fontoura. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. USA 104:1853-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schafer, S. L., R. Lin, P. A. Moore, J. Hiscott, and P. M. Pitha. 1998. Regulation of type I interferon gene expression by interferon regulatory factor-3. J. Biol. Chem. 273:2714-2720. [DOI] [PubMed] [Google Scholar]
- 99.Seo, S. H., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950-954. [DOI] [PubMed] [Google Scholar]
- 100.Seo, S. H., E. Hoffmann, and R. G. Webster. 2004. The NS1 gene of H5N1 influenza viruses circumvents the host anti-viral cytokine responses. Virus Res. 103:107-113. [DOI] [PubMed] [Google Scholar]
- 101.Seo, S. H., and R. G. Webster. 2002. Tumor necrosis factor alpha exerts powerful anti-influenza virus effects in lung epithelial cells. J. Virol. 76:1071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seth, R. B., L. Sun, and Z. J. Chen. 2006. Antiviral innate immunity pathways. Cell Res. 16:141-147. [DOI] [PubMed] [Google Scholar]
- 103.Shin, Y.-K., Q. Liu, S. K. Tikoo, L. A. Babiuk, and Y. Zhou. 2007. Influenza A virus NS1 protein activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by direct interaction with the p85 subunit of PI3K. J. Gen. Virol. 88:13-18. [DOI] [PubMed] [Google Scholar]
- 104.Sims, A. C., R. S. Baric, B. Yount, S. E. Burkett, P. L. Collins, and R. J. Pickles. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 79:15511-15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sims, A. C., B. Yount, S. E. Burkett, R. S. Baric, and R. J. Pickles. 2006. SARS CoV replication and pathogenesis in human airway epithelial cultures. Adv. Exp. Med. Biol. 581:535-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Slepushkin, V. A., P. D. Staber, G. Wang, P. B. McCray, Jr., and B. L. Davidson. 2001. Infection of human airway epithelia with H1N1, H2N2, and H3N2 influenza A virus strains. Mol. Ther. 3:395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Solórzano, A., R. J. Webby, K. M. Lager, B. H. Janke, A. Garcia-Sastre, and J. A. Richt. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stasakova, J., B. Ferko, C. Kittel, S. Sereinig, J. Romanova, H. Katinger, and A. Egorov. 2005. Influenza A mutant viruses with altered NS1 protein function provoke caspase-1 activation in primary human macrophages, resulting in fast apoptosis and release of high levels of interleukins 1β and 18. J. Gen. Virol. 86:185-195. [DOI] [PubMed] [Google Scholar]
- 109.Stewart, W. E., II, W. D. Scott, and S. E. Sulkin. 1969. Relative sensitivities of viruses to different species of interferon. J. Virol. 4:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stonebraker, J. R., D. Wagner, R. W. Lefensty, K. Burns, S. J. Gendler, J. M. Bergelson, R. C. Boucher, W. K. O'Neal, and R. J. Pickles. 2004. Glycocalyx restricts adenoviral vector access to apical receptors expressed on respiratory epithelium in vitro and in vivo: role for tethered mucins as barriers to lumenal infection. J. Virol. 78:13755-13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Szretter, K. J., S. Gangappa, X. Lu, C. Smith, W.-J. Shieh, S. R. Zaki, S. Sambhara, T. M. Tumpey, and J. M. Katz. 2007. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J. Virol. 81:2736-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Takeda, M., A. Pekosz, K. Shuck, L. H. Pinto, and R. A. Lamb. 2002. Influenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J. Virol. 76:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tan, S. L., and M. G. Katze. 1998. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J. Interferon Cytokine Res. 18:757-766. [DOI] [PubMed] [Google Scholar]
- 115.Thanos, D., and T. Maniatis. 1995. Identification of the rel family members required for virus induction of the human beta interferon gene. Mol. Cell. Biol. 15:152-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thanos, D., and T. Maniatis. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83:1091-1100. [DOI] [PubMed] [Google Scholar]
- 117.Thompson, C. I., W. S. Barclay, M. C. Zambon, and R. J. Pickles. 2006. Infection of human airway epithelium by human and avian strains of influenza A virus. J. Virol. 80:8060-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Twu, K. Y., D. L. Noah, P. Rao, R.-L. Kuo, and R. M. Krug. 2006. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J. Virol. 80:3957-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Van Campen, H. 1994. Influenza A virus replication is inhibited by tumor necrosis factor-alpha in vitro. Arch. Virol. 136:439-446. [DOI] [PubMed] [Google Scholar]
- 120.Wan, H., and D. R. Perez. 2007. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J. Virol. 81:5181-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang, G., C. Deering, M. Macke, J. Shao, R. Burns, D. M. Blau, K. V. Holmes, B. L. Davidson, S. Perlman, and P. B. McCray, Jr. 2000. Human coronavirus 229E infects polarized airway epithelia from the apical surface. J. Virol. 74:9234-9239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang, W., and R. M. Krug. 1998. U6atac snRNA, the highly divergent counterpart of U6 snRNA, is the specific target that mediates inhibition of AT-AC splicing by the influenza virus NS1 protein. RNA 4:55-64. [PMC free article] [PubMed] [Google Scholar]
- 123.Wang, W., K. Riedel, P. Lynch, C. Y. Chien, G. T. Montelione, and R. M. Krug. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang, X., C. F. Basler, B. R. G. Williams, R. H. Silverman, P. Palese, and A. Garcia-Sastre. 2002. Functional replacement of the carboxy-terminal two-thirds of the influenza A virus NS1 protein with short heterologous dimerization domains. J. Virol. 76:12951-12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 127.Webster, R. G. 2002. The importance of animal influenza for human disease. Vaccine 20(Suppl. 2):S16-S20. [DOI] [PubMed] [Google Scholar]
- 128.Wong, G. H., and D. V. Goeddel. 1986. Tumour necrosis factors alpha and beta inhibit virus replication and synergize with interferons. Nature 323:819-822. [DOI] [PubMed] [Google Scholar]
- 129.Xu, W., S. Zheng, T. M. Goggans, P. Kiser, M. E. Quinones-Mateu, A. J. Janocha, S. A. A. Comhair, R. Slee, B. R. G. Williams, and S. C. Erzurum. 2006. Cystic fibrosis and normal human airway epithelial cell response to influenza A viral infection. J. Interferon Cytokine Res. 26:609-627. [DOI] [PubMed] [Google Scholar]
- 130.You, Y., E. J. Richer, T. Huang, and S. L. Brody. 2002. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am. J. Physiol. Lung Cell Mol. Physiol. 283:L1315-L1321. [DOI] [PubMed] [Google Scholar]
- 131.Zabner, J., P. Freimuth, A. Puga, A. Fabrega, and M. J. Welsh. 1997. Lack of high affinity fiber receptor activity explains the resistance of ciliated airway epithelia to adenovirus infection. J. Clin. Investig. 100:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zabner, J., B. G. Zeiher, E. Friedman, and M. J. Welsh. 1996. Adenovirus-mediated gene transfer to ciliated airway epithelia requires prolonged incubation time. J. Virol. 70:6994-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zambon, M. C. 2001. The pathogenesis of influenza in humans. Rev. Med. Virol. 11:227-241. [DOI] [PubMed] [Google Scholar]
- 134.Zhang, L., A. Bukreyev, C. I. Thompson, B. Watson, M. E. Peeples, P. L. Collins, and R. J. Pickles. 2005. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 79:1113-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang, L., M. E. Peeples, R. C. Boucher, P. L. Collins, and R. J. Pickles. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 76:5654-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]