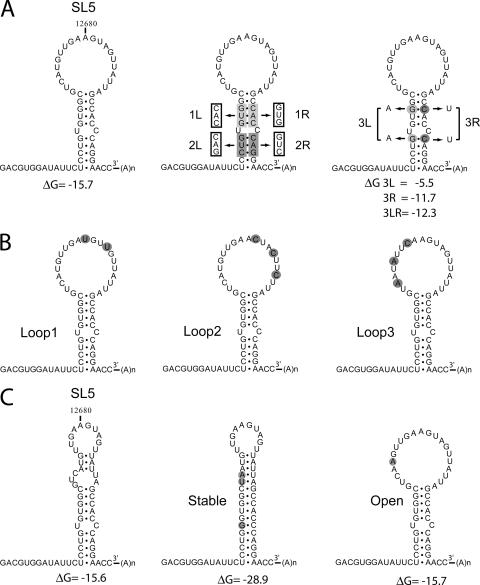
FIG. 1.
RNA secondary structure models for the 3′ UTR of wild-type EAV and SL5 mutants. (A) Mutations were introduced in the SL5 stem region. Mutant 1L has a 3-nucleotide substitution on the left side of the stem, and mutant 1R on the right side. Mutations 1L and 1R are complementary, and base pairing is restored in the double mutant 1LR. Similar mutations were also introduced in the lower stem segment (2L, 2R, and 2LR). The stability of the stem region was targeted in mutants 3L, 3R, and 3LR by the replacement of two G-C base pairs. (B) Mutants Loop1, Loop2, and Loop3 contain point mutations in the SL5 loop region. (C) Mfold prediction of an alternative structure for the SL5 stem-loop with similar stability, in which part of the loop region is closed by base pairing. This alternative structure was stabilized by the mutations introduced in mutant Stable and prevented by the mutation introduced in mutant Open.