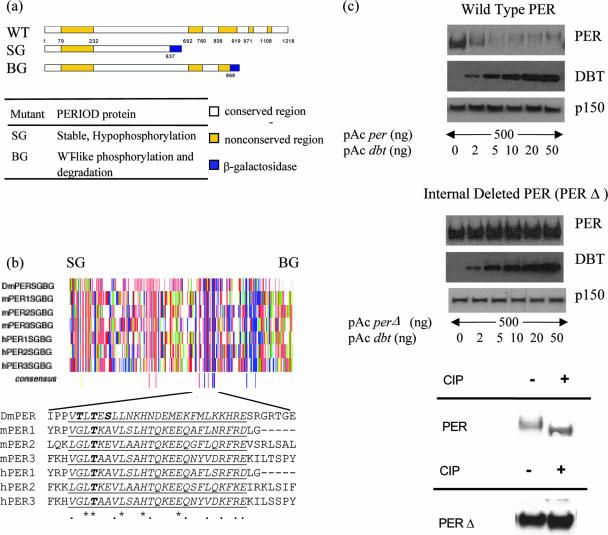
FIG. 1.
Identification of a conserved region in Drosophila PER protein necessary for the progressive phosphorylation. (a) Comparative size of wild-type (WT) and two truncated forms (SG and BG) of PER protein. Numbers represent amino acid positions. (Bottom) Table summarizes behavior of PER-SG and PER-BG in wild-type flies (concluded from references 10 and 47). Conserved and nonconserved regions are as defined by Colot et al. (6). (b) Amino acid sequence alignment of Drosophila and mammalian PER proteins reveals a short highly conserved sequence from aa 637 to 868 in Drosophila PER which is present in PER-BG but not in PER-SG. Each vertical line represents a single amino acid; color represents its chemical property (acidic [D and E] in red, hydrophobic [A, G, I, L and V] in gray, amido [N and Q] in violet, aromatic [F, W and Y] in orange, basic [R, H and K] in blue, hydroxyl [S and T] in pink, proline in green, and sulfur containing [C and M] in yellow; single-letter codes for amino acids are in brackets). SG and BG signify the last amino acids in PER-SG and PER-BG, respectively. The alignment was performed using CLUSTALW in McVector software. Asterisks indicate identical amino acids, and dots indicate similar amino acids. Underlined is a putative conserved sequence. (c) Deletion of the conserved sequence renders Drosophila PER resistant to DBT-induced progressive phosphorylation and degradation (top and middle) and hypophosphorylated (bottom). Western blot of wild-type PER (top) and of PERΔ (from which the conserved sequence is removed) (middle) from S2 cells overexpressing PER and increasing amounts of DBT. PER and DBT were detected using mouse anti-V5 antibody. p150 protein is used as a loading control. CIP, calf intestine phosphatase.