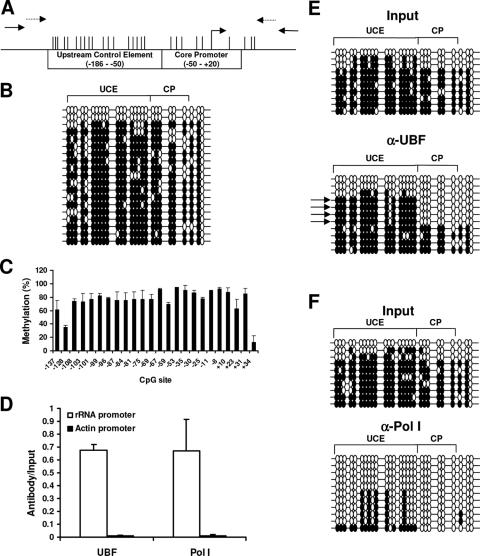
FIG. 1.
The rRNA promoter in HeLa cells is highly methylated; however, the active rRNA promoters bound to Pol I and UBF are unmethylated. (A) Physical map of the rRNA promoter, with positions of the CGs indicated by vertical bars. The primers used for the ChIP assay and bisulfite mapping are indicated with solid and dashed arrows, respectively. (B) Bisulfite analysis of the rRNA promoter from DNA extracted from HeLa cells. Each line represents an independent clone. A filled circle represents a methylated CG dinucleotide, and an empty circle represents an unmethylated CG dinucleotide. (C) Semiquantitative analysis of the C:T peaks of CG dinucleotides as determined through sequencing of the sodium bisulfite PCR product. (D) Chromatin immunoprecipitation analysis of the association between UBF or Pol I binding to the rRNA promoter in HeLa cells. To control for specificity of the antibodies used, the β-ACTIN promoter was amplified for all samples. The graph represents results from quantitative real-time PCR analysis of the rRNA promoter and the β-ACTIN promoter from anti-UBF or anti-Pol I immunoprecipitates normalized to the input samples from three independent experiments. One-tenth of the input sample was used for PCR amplification. (E and F) Bisulfite analysis of rRNA promoters from input samples or from samples immunoprecipitated with anti-UBF (α-UBF) antibody (E) or anti-Pol I antibody (F). The arrows point out the predominant DNA methylation pattern bound to UBF. The results are representative of three independent experiments.