Abstract
Pre-mRNA splicing is essential to ensure accurate expression of many genes in eukaryotic organisms. In Entamoeba histolytica, a deep-branching eukaryote, approximately 30% of the annotated genes are predicted to contain introns; however, the accuracy of these predictions has not been tested. In this study, we mined an expressed sequence tag (EST) library representing 7% of amoebic genes and found evidence supporting splicing of 60% of the testable intron predictions, the majority of which contain a GUUUGU 5′ splice site and a UAG 3′ splice site. Additionally, we identified several splice site misannotations, evidence for the existence of 30 novel introns in previously annotated genes, and identified novel genes through uncovering their spliced ESTs. Finally, we provided molecular evidence for the E. histolytica U2, U4, and U5 snRNAs. These data lay the foundation for further dissection of the role of RNA processing in E. histolytica gene expression.
Eukaryotic genes are often expressed as discontinuous units requiring the removal of intervening RNA sequences (introns) in order to discern their reading frames and ensure their accurate expression. The pre-mRNA-splicing reaction partners are brought into proximity through dynamic rearrangements of the spliceosome, a RNP complex composed of numerous snRNPs and five noncoding snRNAs: U1, U2, U4, U5, and U6 (18, 27). The precise splice sites are characterized by conserved sequence elements.
Entamoeba histolytica infects an estimated 500 million people annually (41). Cysts are ingested in food and water contaminated with fecal matter and excyst into the disease-causing trophozoite in the small intestine. In most people, this results in asymptomatic colonization and reencystation with no subsequent pathology. However, 50 million of those infected each year develop invasive disease (bloody diarrhea or liver abscesses) (41). How E. histolytica regulates gene expression during host invasion, encystation, excystation, and trophozoite vegetative growth is largely unknown.
Prior to completion of the E. histolytica genome sequence, only a few introns had been reported (24, 33, 34, 40). Based on these limited data, the consensus amoebic 5′ and 3′ splice sites (5′, GUUUGU; 3′, UAG) and the lack of a well-conserved branch point consensus were described (40) and incorporated into the computational gene finders used for genome annotation (24). Given that only a few examples of introns had ever previously been uncovered, it was surprising that the genome-sequencing project revealed 3,188 introns in the 9,938 predicted genes (24). Correct intron removal is therefore a necessity for the accurate expression of at least a third of the presently annotated E. histolytica genes. However, the vast majority of these intron predictions lacked molecular validation. The absence of a systematic test of splice site predictions and splicing in this organism presents a significant barrier to our ability to understand its genome structure and the role of RNA processing in amoebic gene regulation.
In this study, we computationally mined an E. histolytica expressed sequence tag (EST) library for hallmarks of splicing. The questions we sought to address were (i) how accurate are the current intron predictions and (ii) how complete is our understanding of splicing in this organism. We compared the intron predictions to the processing patterns deduced from EST analysis, mined the ESTs for novel introns, and used covariance models to computationally identify E. histolytica snRNAs. We found evidence supporting the splicing of several predicted introns and identified several splice site misannotations, novel introns in annotated genes, and novel intron-containing genes. In addition, we identified EST evidence for intron retention and provided molecular evidence for U2, U4, and U5 snRNAs. These data are the result of the largest-scale test of splicing in this organism to date and form the basis for dissecting the interplay between the spliceosome and other cellular machinery involved in amoebic-gene regulation.
MATERIALS AND METHODS
E. histolytica EST library and data sets.
The E. histolytica EST library was created from pooled total RNA (from parasites in the mid-log and stationary phases and from a mouse model of amoebic colitis) (Barbara Mann, personal communication). The datasets containing the intron predictions, EST sequences, and gene predictions were downloaded from The Institute for Genomic Research (http://www.tigr.org/tdb/e2k1/eha1/).
Computational mapping of ESTs to the genome scaffolds.
In order to determine which genomic loci were likely to encode the ESTs, we aligned the EST sequences to the genome sequence data, using the BLAT alignment program (with the version 30 default parameters) (20). Per the default parameters, there were no restrictions on the size of the gap or the amount of 5′ and 3′ overlap between the ESTs and the genomic sequence. Because each EST should be nearly identical to the corresponding genomic region (some mismatch was allowed for sequencing errors), we considered alignments that had ≥98% sequence identity between the genomic regions and the full-length EST transcript (matches of >0.98 × QuerySize).
Computational mining of the EST alignments for introns.
In order to identify possible introns, we computed the coordinates of unaligned gap regions (i.e., the putative introns) from the BLAT alignments described above. The EST gap coordinates were computationally compared to the 3,188 predicted intron coordinates determined from the genome sequence project (24). If the EST gap coordinates matched the coordinates of a predicted intron, we counted this intron as “spliced as predicted” (see Table S1 in the supplemental material). If the EST gap coordinates did not match the predicted intron, we counted that intron as “spliced but at coordinates other than that which are predicted” (Table 1). If the EST gap coordinates did not map to a region known to contain an intron or gene, we deemed that intron “novel.” If no ESTs mapped to a region containing a predicted intron, we deemed that intron prediction “untestable” and did not consider it further. If only ungapped ESTs mapped to a region containing a predicted intron, we deemed that intron “not spliced as predicted” (see Table S2 in the supplemental material).
TABLE 1.
E. histolytica intron splice site misannotationsa
| Representative EST | Transcript | GenBank accession no. | Predicted function | Predicted sequence at indicated splicing site:
|
Predicted coordinates | Predicted size | BLAT-EST sequence at indicated splicing site:
|
BLAT-EST coordinates | BLAT- EST size | No. of unique ESTs | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ | 3′ | 5′ | 3′ | |||||||||
| EHAG374TR | 6.m00476 | ELA51573.1 | Hypothetical protein | GUAUUU | GAG | 122713-123030 | 317 | GUUUGU | UAG | 122735-122797 | 62 | 1 |
| EHAA018TR | 18.m00295 | ELA50710.1 | Hypothetical protein | GUUUAU | AAG | 16364-16517 | 153 | GUUUGU | UAG | 16351-16463 | 112 | 66 |
| EHAAB92TF | 88.m00175 | ELA47893.1 | Hypothetical protein | GUAUUG | GAG | 56505-57006 | 501 | GUUUGU | UAG | 56507-56681 | 174 | 10 |
| EHAE529TR | 50.m00171 | ELA49178.1 | Vesicular transport | GUUGGA | UAG | 22303-22390 | 87 | GUUUGU | UAG | 22293-22368 | 75 | 1 |
| EHAC267TR | 42.m00212 | ELA49475.1 | S6 Ribosomal protein | GUUAUG | UAG | 76818-76880 | 62 | GUAUGU | UAG | 76814-76870 | 56 | 3 |
| EHAC267TR | 42.m00214 | ELA49453.1 | S6 Ribosomal protein | GUUAUG | UAG | 86245-86307 | 62 | GUAUGU | UAG | 86241-86296 | 56 | 3 |
| EHAC267TR | 1066.m00011 | ELA42451.1 | S6 Ribosomal protein | GUCCAU | AAG | 270-365 | 95 | GUAUGU | UAG | 280-335 | 56 | 3 |
| EHAES83TR | 350.m00049 | ELA43647.1 | Rab family GTPase | GUAAUU | UAG | 6630-6679 | 50 | GUUUGU | UAG | 6625-6679 | 54 | 3 |
| EHAES83TR | 350.m00049 | ELA43647.1 | Rab family GTPase | GUUUGU | UAG | 6442-6562 | 121 | GUUUGU | UAG | 6442-6503 | 61 | 3 |
| EHAES83TR | 350.m00049 | ELA43647.1 | Rab family GTPase | GUUUGU | UAG | 6442-6562 | 121 | GUUUGU | UAG | 6513-6563 | 50 | 3 |
Data for eight intron-containing genes for which the intron prediction did not match the EST data are shown.
Computational identification of snRNAs.
We computationally identified the U2 and U4 spliceosomal RNAs using a combination of Hidden Markov models (HMMs) and stochastic context-free grammars (SCFGs), techniques that search for conservation in the primary sequence and secondary structures between a query sequence and a training set (3, 11, 23, 36). U2 and U4 in the Rfam database Release 7.0 were used to train the above programs (14, 15). The majority of the genome sequence was filtered out, using HMMs (default parameters, version 2.3.2). The remainder of the genome sequence with the greatest similarity to known U2 and U4 snRNAs was further scored, using an SCFG (internal package) against the models obtained from Rfam (default parameters, version 0.7). In order to identify U5 snRNA, we downloaded all 235 full sequences of U5 from the Rfam database. We used BLAT (standard parameters, version 30) to align each of these sequences against the full E. histolytica genome sequence.
E. histolytica cell culture, RNA, and DNA isolation.
E. histolytica strain HM-1:IMSS was grown axenically in Trypticase-yeast extract-iron-serum (TYI-S-33) medium (9, 26). Trophozoites were grown to log phase, and total RNA was isolated, using Trizol reagent. Genomic DNA was isolated as indicated by Ali et al. (1).
RT-PCR and Northern blot analysis.
One microgram of total RNA was treated with DNase I and incubated with 0.5 μg of oligo(dT)15 for 10 min at 95°C, and reverse transcription and cDNA amplification were performed as by Ehrenkaufer et al. (12). The PCR products were electrophoresed on a 6% native acrylamide gel and stained with ethidium bromide. The cDNA PCR products were cloned into a TOPO-TA vector (Invitrogen) and sequenced, and splicing of the intron was determined based on its absence from the cDNA. For Northern blot analysis, 10 μg of total RNA from E. histolytica HM-1:IMSS trophozoites was electrophoresed on a 6% acrylamide-7 M urea gel along with a radiolabeled 10-base-pair marker (Invitrogen), transferred onto a Hybond-N+ (Amersham) nylon membrane, and cross-linked, using a Stratalinker. Oligonucleotide probes (see Table S3 in the supplemental material) were prepared and used to probe the membrane as described by Davis and Ares (7).
Nucleotide sequence accession numbers.
The following sequences have been deposited in GenBank under the numbers indicated: U2 snRNA, BK006130; U4 snRNA, BK006131; and U5 snRNA, BK006132.
RESULTS AND DISCUSSION
E. histolytica intron predictions.
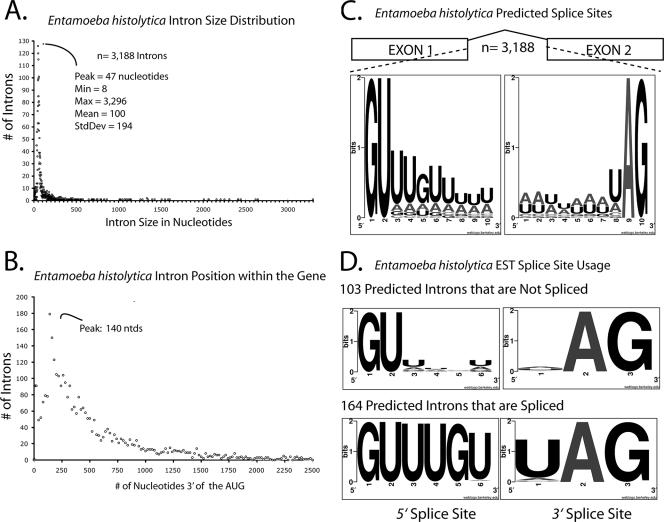
The E. histolytica genome sequence was completed in 2005 and led to a list of 3,188 putative introns in 9,938 predicted genes (24). This is a substantial number of introns compared to the paucity of introns in the related protists Giardia lamblia and Trichomonas vaginalis, suggesting that splicing plays a greater role in amoebic-gene regulation (4, 32, 35, 38). In order to gather a global view of the predicted introns, we determined their sizes and their positions with respect to the start codon and the nucleotide frequencies at the 5′ and 3′ splice donors. Distribution analysis of the predicted E. histolytica intron sizes indicated that the vast majority are small, ∼40 nucleotides in length (Fig. 1A). This is consistent with previous reports of small introns in E. histolytica (33, 39, 40) and comparable to intron sizes from the single-cell parasites T. vaginalis and G. lamblia (4, 32, 35, 38). We noticed that 35 of the predicted E. histolytica introns are smaller than 23 nucleotides. Although spliceosomal introns as small as 23 nucleotides have been validated in the ciliated Paramecium (42), the 23-nucleotide intron size may reflect a lower limit on the geometric constraints for snRNA binding and lariat formation; thus, we concluded that these introns are likely not real (see Table S2 in the supplemental material). Finally, we found that in E. histolytica, the highest proportion of introns are located over the 5′ end of the transcript length (Fig. 1B), a feature commonly found in intron-sparse genomes (29).
FIG. 1.
E. histolytica intron attributes. (A) Histogram showing distribution of 3,188 predicted E. histolytica intron sizes. The majority of introns are ∼40 nucleotides. Note that the x-axis bin size is 1 and that only the major units (50 nucleotides) are depicted. (B) Histogram showing distribution of the distance from the predicted AUG (start codon) to the 5′ splice site for each of the 3,188 predicted introns. Note that the x-axis bin size is 5. (C) Sequence LOGO plot illustrating the relative frequency of nucleotide usage for the first 10 nucleotides (extended 5′ splice site) and the last 10 nucleotides (extended 3′ splice site) for each of the 3,188 predicted introns. (D) Sequence LOGO plot of the first six nucleotides and last three nucleotides of the introns that are spliced as predicted in the ESTs (lower panel) or not spliced as predicted (upper panel).
Analyses of the predicted splice sites indicate that the primary 5′ splice site is composed of GUUUGU and the 3′ splice site is UAG (Fig. 1C), consistent with the previous limited reports of introns in E. histolytica (25, 34, 40). One of the unique features of the spliceosomal introns identified in T. vaginalis and G. lamblia is the incorporation of a well-conserved branch point sequence into an extended 3′ splice site (32, 35). Of the known T. vaginalis introns, the branch point sequence ACUAAC is incorporated into the extended 3′ splice site, prompting speculation that T. vaginalis spliceosomes may combine the steps of branch point- and 3′-splice site recognition (38). In contrast, only 90 of the 3,188 predicted E. histolytica introns contain this sequence (data not shown), indicating that this branch point sequence is not strictly conserved in E. histolytica introns. However, sequences that resemble the degenerate mammalian branch point are found in many E. histolytica introns (40). Lastly, a substantial number of E. histolytica genes are predicted to contain multiple introns (24), raising the issue of whether some of these genes undergo regulated or alternative splicing.
Comparison of intron predictions with EST splice patterns.
Although 3,188 introns have been predicted in E. histolytica, less than 20 have been experimentally validated (25, 33, 40). In order to determine the accuracy of the intron predictions, we directly compared the predicted introns to their spliced counterparts by mining an EST library for hallmarks of splicing. To accommodate the putative intron, we allowed for gaps of ≥23 nucleotides to occur in the EST relative to its genome sequence (Fig. 1A). Of the 3,188 predicted intronic loci, 275 are spanned by ESTs that satisfy these criteria and are therefore testable. In order to determine if the predictions matched the ESTs, we compared the EST gap coordinates to those of the predicted intron. One hundred sixty-four of the EST gap coordinates matched the coordinates of the predicted intron, indicating that they are spliced exactly as annotated (see Table S1 in the supplemental material), at splice sites primarily composed of GUUUGU-UAG (Fig. 1D). However, for other introns, the predicted coordinates did not match those deduced from the ESTs, indicating that these predictions are incorrect (Table 1). In general, we noticed that splice sites that were incorrectly predicted to use a splice donor other than the preferred GUUUGU are not used in vivo, in favor of a nearby GUUUGU. Likewise, a nearby UAG 3′ splice acceptor site appears to be utilized over GAG, AAG, and, in some instances, even a neighboring UAG. Moreover, in nearly all cases, the spliced intron was smaller than predicted. Lastly, although 103 of the 275 testable putative introns contain canonical splice sites, we failed to find evidence for their removal in any of their corresponding ESTs (Fig. 1D; also see Table S2 in the supplemental material). This suggests that either these are not introns, are not spliced under conditions represented in the EST library, or have such low splicing efficiency that no spliced isoforms were cloned.
Mining the ESTs for unannotated introns and genes.
In order to identify novel processing events within the E. histolytica EST database, we mined the ESTs for transcripts with intron-like features independent of any prior predictions. We queried the ESTs for regions that have two or more blocks of sequence with at least 98% identity to the genomic sequence and are separated by a gap of 40 to 200 nucleotides and hand collated the data. In total, we identified 35 novel introns, each of which was classified into one of three categories based on how it affected the protein-reading frame (Table 2).
TABLE 2.
Novel E. histolytica introns culled from the EST dataa
| Representative EST | Transcript | GenBank accession no. | Predicted function | 5′ Splicing site | 3′ Splicing site | BLAT coordinates | BLAT scaffold | Size | No. of unique ESTs | Effect on protein |
|---|---|---|---|---|---|---|---|---|---|---|
| Class I | ||||||||||
| EHAA254TF | 101.m00114 | ELA47515.1 | Conserved hypothetical protein | GUUUGU | AAG | 10427-10489 | 101 | 62 | 1 | Alters the C terminus |
| EHAA741TF | 42.m00181 | N/A | Pseudogene Ras family GTPase | GUUUGU | UAG | 30857-30960 | 42 | 103 | 7 | Alters the C terminus |
| EHAF244TR | 19.m00316 | ELA50600.1 | Hypothetical protein | GUUUGU | UAG | 77147-77207 | 19 | 60 | 1 | Eliminates amino acids |
| EHAA453TR | 178.m00101 | ELA45803.1 | 3′ UTR of hypothetical protein | GUUUGU | AAG | 36163-36214 | 178 | 51 | 3 | Alters the C terminus |
| EHAA547TR | 110.m00129 | ELA47260.1 | Rho family GTPase | GUUUGU | UAG | 38325-38392 | 110 | 67 | 2 | Alters the C terminus |
| EHABP41TR | 254.m00073 | ELA44597.1 | Hypothetical protein | GUUUGU | AAG | 5957-6014 | 254 | 57 | 3 | Alters the C terminus |
| EHADQ25TR | 18.m00335 | ELA50675.1 | DNA replication licensing factor | GUUUGU | UAG | 98259-98343 | 18 | 84 | 1 | Alters the C terminus |
| EHAET77TR | 18.m00328 | ELA50668.1 | Molybdopterin biosynthesis | GUUUGU | UAG | 90251-90312 | 18 | 61 | 2 | Alters the C terminus |
| EHAGK16TR | 264.m00090 | ELA44495.1 | Sec13 protein | GUAUGU | UAG | 5564-5620 | 264 | 56 | 3 | Alters the N terminus |
| EHAH331TR | 264.m00090 | ELA44495.1 | Sec13 protein | GUUUGU | UAG | 5656-5710 | 264 | 54 | 2 | Alters the N terminus |
| EHAE226TR | 133.m00132 | N/A | Hypothetical protein | GUUUGU | UAG | 16335-16395 | 133 | 60 | 1 | Alters the N and C termini |
| EHAB255TR | 52.m00167 | ELA49102.1 | Rho family GTPase | GUUUGU | UAG | 69361-69424 | 52 | 63 | 5 | Alters the C terminus |
| EHAA702TR | 231.m00059 | ELA44885.1 | Conserved hypothetical protein | GUUUGU | AAG | 7731-7834 | 231 | 103 | 8 | Alters the N terminus |
| EHAG185TRb | 47.m00184 | ELA49297.1 | Hypothetical protein | GUUUGU | AAG | 70138-70195 | 47 | 57 | 1 | Alters the N terminus |
| EHABT01TR | 57.m00155 | ELA48912.1 | Hypothetical protein | GUUCGU | UAG | 34651-34702 | 57 | 51 | 1 | Eliminates amino acids |
| EHAE044TR | 364.m00046 | ELA43561.1 | 60S ribosomal protein L27a | GUUUGU | UAG | 15314-15429 | 364 | 115 | 1 | Alters the N terminus |
| EHAET36TR | 135.m00095 | ELA46630.1 | Hypothetical protein | GUUUGU | UAG | 9119-9259 | 135 | 140 | 6 | Alters the N terminus |
| EHADY14TR | 366.m00044 | ELA43555.1 | Hypothetical protein | GUUUGU | UAG | 6738-6786 | 366 | 48 | 1 | Alters the N terminus |
| EHAG990TR | 152.m00113 | ELA46298.1 | Hypothetical protein | GUUUGU | UAG | 24791-24927 | 152 | 136 | 1 | Alters the N terminus |
| EHAAP93TR | 195.m00094 | ELA45475.1 | 60S ribosomal protein L24 | GUUUGU | UAG | 31405-31461 | 195 | 56 | 2 | Alters the N terminus |
| Class II | ||||||||||
| EHAAY54TR | 88.m00175 | ELA47893.1 | 3′ UTR of hypothetical protein | GUUUGU | UAG | 55917-56020 | 88 | 103 | 2 | N/A |
| EHAE226TR | 23.m00311 | ELA50387.1 | 3′ UTR of in Rho GTPase | GUUUGU | UAG | 24514-24574 | 23 | 60 | 1 | N/A |
| EHAES83TR | 350.m00049 | ELA43647.1 | 5′ UTR of in Rho GTPase | GUUAAG | UAG | 5995-6143 | 350 | 148 | 1 | N/A |
| EHAA378TRb | 21.m00231 | ELA50513.1 | 5′ UTR of 40S ribosomal protein S14 | GUUUGU | UAG | 17466-17539 | 21 | 73 | 3 | N/A |
| EHAA726TR | 312.m00035 | ELA43981.1 | 5′ UTR of 60S ribosomal protein L9 | GUUUGU | UAG | 6822-6956 | 312 | 134 | 12 | N/A |
| EHAHG49TR | 144.m00101 | ELA46471.1 | Similar to cap binding protein | GUUUGU | UAG | 16107-16176 | 144 | 69 | 1 | N/A |
| EHAF084TR | 39.m00252 | ELA49583.1 | Glycotransferase | GUUUGA | UAG | 80547-80602 | 39 | 55 | 1 | N/A |
| Class III | ||||||||||
| EHAAM93TR | N/A | N/A | Similar to 6.m00429 | GUUUGA | UAG | 14120-14172 | 338 | 52 | 1 | New to E. histolytica |
| EHACJ50TRb | N/A | N/A | Similar to pantothenate kinase | GUUUGU | AAG | 76031-76107 | 39 | 76 | 1 | New to E. histolytica |
| EHAFD09TR | N/A | N/A | Similar to UFD1-1 | GUUUGU | UAG | 73812-73859 | 11 | 47 | 1 | New to E. histolytica |
| EHAC353TRb | N/A | N/A | Similar to acriflavin resistance protein | GUUUGU | UAG | 4398-4451 | 389 | 53 | 2 | New to E. histolytica |
| EHAEL21TR | N/A | N/A | Similar to YIP1 Golgi protein | GUUUGU | UAG | 52191-52240 | 62 | 49 | 2 | New to E. histolytica |
| EHAEU30TR | N/A | N/A | CCCH-domain protein | GUUAGU | UAG | 89669-89734 | 5 | 65 | 2 | New to E. histolytica |
| EHACJ32TR | N/A | N/A | CCCH domain protein | GUUUGU | UAG | 90154-90220 | 5 | 66 | 2 | New to E. histolytica |
| EHAHB45TRb | N/A | N/A | No homology, novel | GUUUGU | UAG | 23491-23547 | 154 | 56 | 1 | New to E. histolytica |
Class I introns align by BLAT to genes that were not annotated to contain an intron in that region. Class II introns align by BLAT to the UTRs of genes. Class III introns align by BLAT to regions that were not annotated to contain genes. N/A, not applicable.
EST for which the spliced product has been cloned and sequenced.
Class I introns.
Class I is the largest class of novel introns we identified. These introns are located in or near annotated genes but in regions not annotated to be intronic; i.e., they were predicted to be exonic or in regions immediately proximal to an open reading frame. However, in silico translation of the surrounding spliced sequence revealed an extension of the protein-coding region of the adjacent genes.
Class II introns.
Class II introns map immediately proximal to annotated open reading frames. However, in contrast to Class I introns, in silico translation of the surrounding spliced sequences did not alter the protein-coding region of the adjacent genes, suggesting that these introns reside in their untranslated regions (UTRs). Thus, their retention or removal does not affect the protein-coding potential of the gene.
Class III introns.
Class III introns are located in regions currently annotated as “intergenic” and not predicted to have any protein-coding potential. However, in silico translation of the spliced sequences surrounding the introns uncovered several novel proteins with extended reading frames. Most of these predicted genes have not been previously identified in E. histolytica but have homologs in other organisms. One of the novel genes (on BLAT scaffold 154) lacks homology to any known proteins and contains two introns (one represented by an EST and the other identified computationally while deciphering the protein-reading frame). Splicing of both introns was confirmed by reverse transcription (RT)-PCR and cDNA sequencing (data not shown).
RT-PCR validation and sequencing of the BLAT intron predictions.
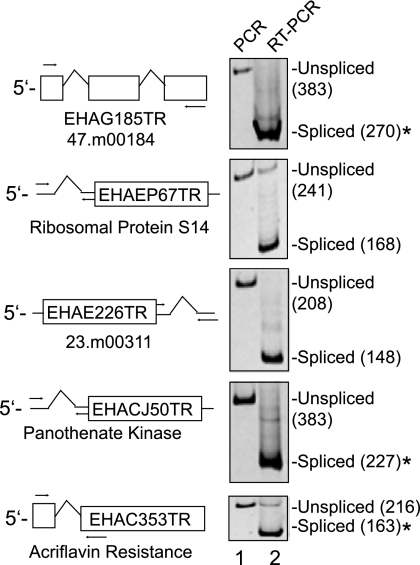
In order to experimentally confirm splicing of the novel introns identified above, we performed RT-PCR on cDNA generated from log-phase E. histolytica HM-1:IMSS trophozoites grown under standard axenic culture conditions. In all cases tested, PCR amplification of cDNA using exonic primers spanning the novel introns generated a product smaller than that amplified from genomic DNA, consistent in size with that from splicing of the predicted introns from these transcripts (Fig. 2). The cDNAs for acriflavin resistance protein, pantothenate kinase, 47.m00184, 21.m00231, and (154.m), a novel gene with no homology to any known protein in the GenBank database, were cloned and sequenced (data not shown). In all cases, the sequencing results confirmed that the splice sites indicated in Table 2 were used. Given the canonical splice donor and acceptor sequences in Table 2, we expect that these remaining novel introns are likewise correct. These data demonstrate that the novel introns we identified are efficiently spliced in log-phase E. histolytica trophozoites and suggest that many additional introns remain to be uncovered.
FIG. 2.
RT-PCR test of BLAT predictions for E. histolytica introns. PCR amplification from either genomic DNA (lane 1) or oligo(dT)-primed cDNA (lane 2) from RNA of log-phase axenic trophozoites for five of the selected novel introns is shown. A diagrammatic gene model is depicted to the left of the gel wherein a box corresponds to an exon. A caret (^) corresponds to an intron. The designations in the boxes beginning with EH and ending with TR are the names of representative ESTs exhibiting the indicated spliced patterns. The common gene name is also indicated below each gene model. Arrows represent the relative positions of the PCR primers. The products were run on a 6% native acrylamide gel and stained with ethidium bromide. The PCR product sizes are indicated in parentheses, and those marked by an asterisk (*) were cloned and sequenced.
EST evidence for intron retention and alternate 3′-splice site selection.
Multi-intron-containing genes are generally a feature of higher eukaryotes and are often accompanied by alternative splicing, such as exon skipping and mutually exclusive exons (17). Approximately 6% of the presently annotated genes in E. histolytica are predicted to be multi-intron containing (24). However, none of the ESTs that span two or more predicted introns in a gene exhibit evidence for exon skipping and mutually exclusive exons (data not shown). Moreover, we found no evidence of exon skipping or mutually exclusive exons in RT-PCR experiments in log-phase E. histolytica trophozoites using primers that span several exons in 10 other multi-intron-containing genes (data not shown).
Other forms of alternative splicing, such as intron retention, are more prevalent in lower eukaryotes with fewer multi-intron-containing genes and smaller introns (21). In order to see if there was any evidence in the ESTs for intron retention, we sought to compare the number of spliced ESTs to the number of unspliced ESTs for each of the 164 introns for which there is functional/EST evidence of splicing (see Table S1 in the supplemental material). While 87% of the 164 introns are spliced in 100% of their representative ESTs, 13% are spliced in only a fraction of their representative ESTs. Two possibilities can readily explain this observation: (i) the fraction of “unspliced” ESTs for an individual intron are derived from its pre-mRNAs cloned prior to splicing; or (ii) the fraction of “unspliced” ESTs for an individual intron are derived from a distinct growth condition in which the intron is selectively retained, i.e., intron retention. Additional directed and high-throughput experiments, such as splicing-sensitive microarray (5), and larger cDNA libraries are needed to identify individual processing events and monitor the alterations in processing during parasite growth and development.
Examples of regulated splicing have been described in other systems as a mechanism to turn transcripts on and off (2, 6, 8, 22, 37). Because we have not tested every growth condition in the life of an amoeba, we cannot formally exclude the possibility that the 37% of introns for which we see no evidence of splicing are indeed spliced under a given condition. One point at which alternate isoforms of the same pre-mRNA may be generated is the developmental switch between the trophozoite and cyst forms of E. histolytica. Microarray data indicate that ∼15% of annotated genes change ±3-fold between trophozoites and cysts of E. histolytica (12). Whether these changes in RNA abundance between the life cycle stages reflect alterations in transcription frequency or decay as a result of regulated processing remains to be tested.
Finally, some genes are known to generate different proteins as a result of splicing at alternate 5′ and 3′ splice sites (10, 16). In order to see if there was any evidence in the EST library for alternate 5′- and 3′-splice site usage, we individually mined each spliced intron for examples of ESTs in which all of the coordinates for one of the splice sites was fixed while the other varied. We found no evidence for alternate 5′-splice site usage. However, 89.m00113, a gene with similarity to human Sm_B/B′ protein, has representative ESTs in which different 3′ splice sites are used for the penultimate intron, which would introduce two additional amino acids in the C terminus (data not shown). Curiously, the human Sm_B and Sm_B′ isoforms are derived from alternative splicing using different 3′ splice sites of the penultimate intron that are distinguishable by autoantibodies generated in people with systemic lupus erythematosus (19). Thus, overall, we found EST evidence for candidate intron retention and alternative 3′-splice site usage.
E. histolytica spliceosomal RNAs (snRNAs).
snRNAs bound in the spliceosomal complex of over 150 proteins interact with the intron through RNA-RNA interactions (18). The pre-mRNA reaction partners for the two catalytic steps of splicing are brought into proximity through dynamic rearrangements of the pre-mRNA/snRNA and snRNA/snRNA complexes requiring U1, U2, U4, U5, and U6 snRNAs (27). To date, U6 is the only E. histolytica snRNA that has been identified (28). Given the essential role of the snRNAs in splicing, we queried the E. histolytica genome for the presence of the U1, U2, U4, and U5 snRNAs.
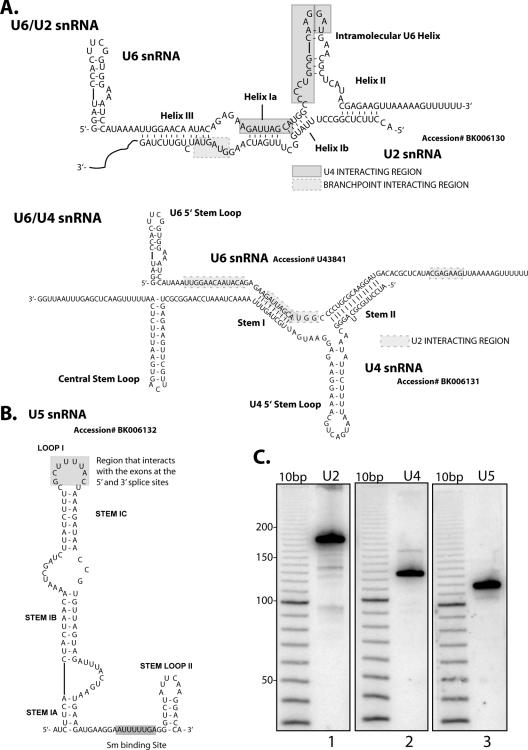
U2 snRNA is involved in pre-mRNA/snRNA base pairing and juxtapositioning of the branch point adenosine for the first transesterification reaction. In order to identify the E. histolytica U2 snRNA, we downloaded 553 U2 snRNA sequences from Rfam and built an HMM to look for conserved features. The region on scaffold 25 from 23993 to 24173 had the greatest similarity to known U2 snRNAs and was selected for Northern blot analysis. We saw U2 accumulate as a predominate species, 178 nucleotides in length, in trophozoite RNA (Fig. 3C). Its putative secondary structure is similar to those of other known U2 snRNAs, including the branch point binding sequence and the Sm binding site (data not shown), and is predicted to interact with U6 snRNA in the conserved fashion. The U4 snRNA base pairs with U6 snRNA, acting as its chaperone and maintaining it in an unfolded conformation while part of the U4/U5/U6 tri-snRNP (13). We applied the above approach to identify U4 snRNA based on the 372 U4 snRNA sequences in Rfam. We identified the region on scaffold 150 from 39898 to 40028. Subsequent Northern blot analysis of this region uncovered a predominant band 125 nucleotides in length (Fig. 3C). This putative U4 snRNA is able to interact with the previously identified U6 snRNA in a conserved fashion. Of note, the U4 snRNA also seems to lack the terminal 3′ stem loop found in higher eukaryotes (30).
FIG. 3.
E. histolytica spliceosomal RNAs (snRNAs). (A) Predicted secondary structures of U2 snRNA bound to U6 snRNA and U4 snRNA bound to U6 snRNA. (B) Predicted secondary structure of U5 snRNA. (C) Northern blots for U2, U4, and U5 snRNAs. Ten micrograms of HM-1:IMSS total RNA was fractioned on a 6% denaturing acrylamide gel and probed with a radiolabeled oligo targeting each of the predicted snRNAs. A radiolabeled 10-base-pair marker (Invitrogen) was loaded in parallel to assess the sizes of each of the snRNAs.
U5 snRNA interacts with the exons upstream of the 5′ splice site and downstream of the 3′ splice site, tethering them in the active site for the second transesterification (31). Our efforts to identify the E. histolytica U5 snRNA using the above means failed. Therefore, we used BLAT for each of the 235 U5 sequences in the Rfam database against the E. histolytica genome scaffolds. We identified a region on scaffold 283 from 9300 to 9468 with significant homology to the U5 sequences from Entosiphon sulcatum, Oryza sativa, Zea mays, and Arabidopsis thaliana. Northern blot analysis of this region uncovered a single band 118 nucleotides in length (Fig. 3C). Secondary structure prediction showed its potential to form the evolutionarily conserved site in stems I and II as well as the Sm binding site (Fig. 3B). Using the computational approaches outlined above, we were unable to identify U1. Whether this indicates that the E. histolytica U1 sequence is substantially different or that it has escaped being sequenced is not clear at present.
Conclusions.
Despite the ability of RNA processing to markedly alter the coding potential of genes, the mechanisms that control these events in E. histolytica are poorly understood. We compared the splice patterns mined from EST data to 275 computational intron predictions. We found evidence supporting the splicing of 60% of introns exactly as predicted. Additionally, we identified several splice site misannotations, novel introns in annotated genes, and novel intron-containing genes. Since the EST data we analyzed represented ∼7% of the predicted amoebic genes, our work indicates that a larger-scale EST library would significantly improve gene annotation and uncover additional useful information regarding mechanisms of RNA processing in E. histolytica. This work represents the first large-scale test of splicing in a deep-branching eukaryote and indicates that similar analyses in other systems may be similarly fruitful.
Supplementary Material
Acknowledgments
We thank all members of the Singh lab, specifically Gretchen Ehrenkaufer and Jason Hackney, for critical and editorial comments on the manuscript; Neil Hall and Lis Caler (TIGR) for providing the EST sequences and incorporating data into genome reannotation; Barbara Mann (University of Virginia) for providing information on the EST library; and Neha Gupta for preliminary RT-PCR analysis of intron-containing genes.
This work was supported by NIH grants AI-053724 to Upinder Singh and T32 AI-07502 to Carrie A. Davis.
Footnotes
Published ahead of print on 27 April 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Ali, I. K., M. Zaki, and C. G. Clark. 2005. Use of PCR amplification of tRNA gene-linked short tandem repeats for genotyping Entamoeba histolytica. J. Clin. Microbiol. 43:5842-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Averbeck, N., S. Sunder, N. Sample, J. A. Wise, and J. Leatherwood. 2005. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol. Cell 18:491-498. [DOI] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, R. D. Finn, and E. L. Sonnhammer. 1999. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 27:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlton, J. M., R. P. Hirt, J. C. Silva, A. L. Delcher, M. Schatz, Q. Zhao, J. R. Wortman, S. L. Bidwell, U. C. Alsmark, S. Besteiro, T. Sicheritz-Ponten, C. J. Noel, J. B. Dacks, P. G. Foster, C. Simillion, Y. Van de Peer, D. Miranda-Saavedra, G. J. Barton, G. D. Westrop, S. Muller, D. Dessi, P. L. Fiori, Q. Ren, I. Paulsen, H. Zhang, F. D. Bastida-Corcuera, A. Simoes-Barbosa, M. T. Brown, R. D. Hayes, M. Mukherjee, C. Y. Okumura, R. Schneider, A. J. Smith, S. Vanacova, M. Villalvazo, B. J. Haas, M. Pertea, T. V. Feldblyum, T. R. Utterback, C. L. Shu, K. Osoegawa, P. J. de Jong, I. Hrdy, L. Horvathova, Z. Zubacova, P. Dolezal, S. B. Malik, J. M. Logsdon, Jr., K. Henze, A. Gupta, C. C. Wang, R. L. Dunne, J. A. Upcroft, P. Upcroft, O. White, S. L. Salzberg, P. Tang, C. H. Chiu, Y. S. Lee, T. M. Embley, G. H. Coombs, J. C. Mottram, J. Tachezy, C. M. Fraser-Liggett, and P. J. Johnson. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, T. A., C. W. Sugnet, and M. Ares, Jr. 2002. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296:907-910. [DOI] [PubMed] [Google Scholar]
- 6.Dabeva, M. D., M. A. Post-Beittenmiller, and J. R. Warner. 1986. Autogenous regulation of splicing of the transcript of a yeast ribosomal protein gene. Proc. Natl. Acad. Sci. USA 83:5854-5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, C. A., and M. Ares, Jr. 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103:3262-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, C. A., L. Grate, M. Spingola, and M. Ares, Jr. 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 28:1700-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamond, L. S., C. G. Clark, and C. C. Cunnick. 1995. YI-S, a casein-free medium for axenic cultivation of Entamoeba histolytica, related Entamoeba, Giardia intestinalis and Trichomonas vaginalis. J. Eukaryot. Microbiol. 42:277-278. [DOI] [PubMed] [Google Scholar]
- 10.Dou, Y., K. L. Fox-Walsh, P. F. Baldi, and K. J. Hertel. 2006. Genomic splice-site analysis reveals frequent alternative splicing close to the dominant splice site. RNA (New York) 12:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eddy, S. R., and R. Durbin. 1994. RNA sequence analysis using covariance models. Nucleic Acids Res. 22:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrenkaufer, G. M., R. Haque, J. A. Hackney, D. J. Eichinger, and U. Singh. 2007. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell. Microbiol. [DOI] [PubMed]
- 13.Gmeiner, W. H. 2002. The structure and dynamics of the U4/U6 snRNP: implications for pre-mRNA splicing and use as a model system to investigate the RNA-mediated effects of (5F)Ura. J. Biomol. Struct. Dyn. 19:853-862. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths-Jones, S., A. Bateman, M. Marshall, A. Khanna, and S. R. Eddy. 2003. Rfam: an RNA family database. Nucleic Acids Res. 31:439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths-Jones, S., S. Moxon, M. Marshall, A. Khanna, S. R. Eddy, and A. Bateman. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33:D121-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiller, M., K. Huse, K. Szafranski, N. Jahn, J. Hampe, S. Schreiber, R. Backofen, and M. Platzer. 2004. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat. Genet. 36:1255-1257. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J. M., J. Castle, P. Garrett-Engele, Z. Kan, P. M. Loerch, C. D. Armour, R. Santos, E. E. Schadt, R. Stoughton, and D. D. Shoemaker. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302:2141-2144. [DOI] [PubMed] [Google Scholar]
- 18.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 19.Kaufman, K. M., M. Y. Kirby, M. T. McClain, J. B. Harley, and J. A. James. 2001. Lupus autoantibodies recognize the product of an alternative open reading frame of SmB/B′. Biochem. Biophys. Res. Commun. 285:1206-1212. [DOI] [PubMed] [Google Scholar]
- 20.Kent, W. J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, E., A. Magen, and G. Ast. 2007. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 35:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kishida, M., T. Nagai, Y. Nakaseko, and C. Shimoda. 1994. Meiosis-dependent mRNA splicing of the fission yeast Schizosaccharomyces pombe mes1+ gene. Curr. Genet. 25:497-503. [DOI] [PubMed] [Google Scholar]
- 23.Krogh, A., M. Brown, I. S. Mian, K. Sjolander, and D. Haussler. 1994. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 235:1501-1531. [DOI] [PubMed] [Google Scholar]
- 24.Loftus, B., I. Anderson, R. Davies, U. C. Alsmark, J. Samuelson, P. Amedeo, P. Roncaglia, M. Berriman, R. P. Hirt, B. J. Mann, T. Nozaki, B. Suh, M. Pop, M. Duchene, J. Ackers, E. Tannich, M. Leippe, M. Hofer, I. Bruchhaus, U. Willhoeft, A. Bhattacharya, T. Chillingworth, C. Churcher, Z. Hance, B. Harris, D. Harris, K. Jagels, S. Moule, K. Mungall, D. Ormond, R. Squares, S. Whitehead, M. A. Quail, E. Rabbinowitsch, H. Norbertczak, C. Price, Z. Wang, N. Guillen, C. Gilchrist, S. E. Stroup, S. Bhattacharya, A. Lohia, P. G. Foster, T. Sicheritz-Ponten, C. Weber, U. Singh, C. Mukherjee, N. M. El-Sayed, W. A. Petri, Jr., C. G. Clark, T. M. Embley, B. Barrell, C. M. Fraser, and N. Hall. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865-868. [DOI] [PubMed] [Google Scholar]
- 25.Lohia, A., and J. Samuelson. 1993. Cloning of the Eh cdc2 gene from Entamoeba histolytica encoding a protein kinase p34cdc2 homologue. Gene 127:203-207. [DOI] [PubMed] [Google Scholar]
- 26.MacFarlane, R. C., and U. Singh. 2006. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis. Infect. Immun. 74:340-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madhani, H. D., and C. Guthrie. 1994. Dynamic RNA-RNA interactions in the spliceosome. Annu. Rev. Genet. 28:1-26. [DOI] [PubMed] [Google Scholar]
- 28.Miranda, R., L. M. Salgado, R. Sanchez-Lopez, A. Alagon, and P. M. Lizardi. 1996. Identification and analysis of the u6 small nuclear RNA gene from Entamoeba histolytica. Gene 180:37-42. [DOI] [PubMed] [Google Scholar]
- 29.Mourier, T., and D. C. Jeffares. 2003. Eukaryotic intron loss. Science 300:1393. [DOI] [PubMed] [Google Scholar]
- 30.Myslinski, E., and C. Branlant. 1991. A phylogenetic study of U4 snRNA reveals the existence of an evolutionarily conserved secondary structure corresponding to ‘free’ U4 snRNA. Biochimie 73:17-28. [DOI] [PubMed] [Google Scholar]
- 31.Newman, A. J., and C. Norman. 1992. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68:743-754. [DOI] [PubMed] [Google Scholar]
- 32.Nixon, J. E., A. Wang, H. G. Morrison, A. G. McArthur, M. L. Sogin, B. J. Loftus, and J. Samuelson. 2002. A spliceosomal intron in Giardia lamblia. Proc. Natl. Acad. Sci. USA 99:3701-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plaimauer, B., S. Ortner, G. Wiedermann, O. Scheiner, and M. Duchene. 1994. An intron-containing gene coding for a novel 39-kilodalton antigen of Entamoeba histolytica. Mol. Biochem. Parasitol. 66:181-185. [DOI] [PubMed] [Google Scholar]
- 34.Roy, S. W., M. Irimia, and D. Penny. 2006. Very little intron gain in Entamoeba histolytica genes laterally transferred from prokaryotes. Mol. Biol. Evol. 23:1824-1827. [DOI] [PubMed] [Google Scholar]
- 35.Russell, A. G., T. E. Shutt, R. F. Watkins, and M. W. Gray. 2005. An ancient spliceosomal intron in the ribosomal protein L7a gene (Rpl7a) of Giardia lamblia. BMC Evol. Biol. 5:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakakibara, Y., M. Brown, R. Hughey, I. S. Mian, K. Sjolander, R. C. Underwood, and D. Haussler. 1994. Stochastic context-free grammars for tRNA modeling. Nucleic Acids Res. 22:5112-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spingola, M., and M. Ares, Jr. 2000. A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol. Cell 6:329-338. [DOI] [PubMed] [Google Scholar]
- 38.Vanácová, S., W. Yan, J. M. Carlton, and P. J. Johnson. 2005. Spliceosomal introns in the deep-branching eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 102:4430-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogel, G. 2006. Infectious diseases. Tackling neglected diseases could offer more bang for the buck. Science 311:592-593. [DOI] [PubMed] [Google Scholar]
- 40.Wilihoeft, U., E. Campos-Gongora, S. Touzni, I. Bruchhaus, and E. Tannich. 2001. Introns of Entamoeba histolytica and Entamoeba dispar. Protist 152:149-156. [DOI] [PubMed] [Google Scholar]
- 41.World Health Organization. 1997. A consultation with experts on amoebiasis. WHO/PAHO/UNESCO report. Mexico City, Mexico, 28-29 January, 1997. Epidemiol. Bull. 18:13-14. [PubMed] [Google Scholar]
- 42.Yamauchi, K., T. Ochiai, and I. Usuki. 1992. The unique structure of the Paramecium caudatum hemoglobin gene: the presence of one intron in the middle of the coding region. Biochim. Biophys. Acta 1171:81-87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.