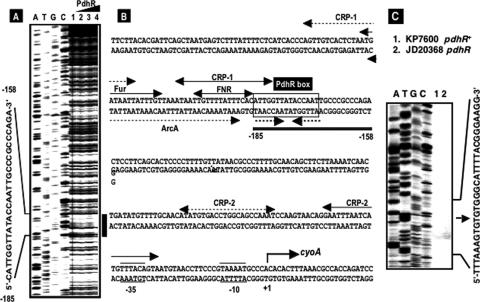
FIG. 4.
DNase I footprinting of the cyoA promoter. (A) The fluorescently labeled cyoA promoter was incubated with increasing amounts of purified PdhR (lane 1, 0 pmol; lane 2, 2.5 pmol; lane 3, 5 pmol; lane 4, 10 pmol) and subjected to DNase I footprinting assays. Lanes A, T, G, and C represent sequence ladders. (B) The black bar (positions −185 to −158) on the right indicates the PdhR-binding region located upstream of the cyoA promoter. The nucleotide numbers represent the distance from the transcription initiation site, which was proposed previously by Minagawa et al. (22) and confirmed in this study. Recognition sequences for Fur, ArcA, and Fnr were predicted based on sequence analyses reported previously by Stojiljkovic et al. (35), Shalel-Levanon et al. (29), and Salmon et al. (28), respectively. Two CRP-binding sequences, CRP1 and CRP2, were proposed previously, but at different positions, by Minagawa et al. (22) (dotted lines) and Zheng et al. (40) (solid line). (C) Primer extension assay of the transcription initiation site of the cyoA operon in both wild-type KP7600 and its pdhR disruptant, JD20368, was performed using a fluorescently labeled probe. Data for the site identified thus agreed with those reported previously by Minagawa et al. (22).