Abstract
Quorum sensing in Vibrio cholerae involves signaling between two-component sensor protein kinases and the response regulator LuxO to control the expression of the master regulator HapR. HapR, in turn, plays a central role in regulating a number of important processes, such as virulence gene expression and biofilm formation. We have determined the crystal structure of HapR to 2.2-Å resolution. Its structure reveals a dimeric, two-domain molecule with an all-helical structure that is strongly conserved with members of the TetR family of transcriptional regulators. The N-terminal DNA-binding domain contains a helix-turn-helix DNA-binding motif and alteration of certain residues in this domain completely abolishes the ability of HapR to bind to DNA, alleviating repression of both virulence gene expression and biofilm formation. The C-terminal dimerization domain contains a unique solvent accessible tunnel connected to an amphipathic cavity, which by analogy with other TetR regulators, may serve as a binding pocket for an as-yet-unidentified ligand.
The gram-negative bacterium Vibrio cholerae is the causative agent of the frequently fatal epidemic disease cholera. The ability of V. cholerae to infect humans is dependent on the expression of virulence factors including the toxin coregulated pilus (48), a critical colonization factor, and cholera toxin. The expression of these genes is dependent upon a transcriptional cascade that is initiated at the tcpPH promoter by the regulators AphA and AphB (26, 45). AphA, which is a member of a new and relatively uncharacterized regulatory family, activates tcpPH expression by facilitating the binding of the LysR-type regulator, AphB, to the promoter (23). The crystal structure of AphA, which has recently been reported by our group, revealed that it is a winged helix DNA-binding protein with an unusual C-terminal antiparallel coiled-coil dimerization domain (7).
Quorum sensing is a process of bacterial cell-cell communication that influences gene expression in a cell density-dependent manner (34). Both gram-negative and gram-positive bacteria use quorum sensing to regulate several physiological functions, including bioluminescence, virulence factor expression, antibiotic production, and biofilm development (34). In most gram-negative bacteria, the quorum-sensing circuit resembles that of the symbiotic bacterium Vibrio fischeri (12), where a LuxI-type protein synthesizes a specific acylated homoserine lactone signaling molecule known as autoinducer. As the cell density increases, the concentration of autoinducer increases and, upon reaching a critical threshold concentration both intra- and extracellularly, it binds to a soluble LuxR-type protein that then activates gene expression. Other examples of this circuit include the TraI/TraR system of Agrobacterium tumefaciens (40, 50), and the LasI/LasR system of Pseudomonas aeruginosa (5).
The quorum-sensing circuit of V. cholerae is significantly more complex than that of the V. fischeri-type circuit and is similar to that of the free-living marine bacterium Vibrio harveyi. At least two different autoinducers, CAI-1 and AI-2, are synthesized by the enzymes CqsA and LuxS, respectively, which share no homology to the LuxI family of autoinducer synthases (35). As the cell density increases, these autoinducers are detected by the cognate sensors CqsS and LuxPQ, respectively, which are hybrid, two-component sensor protein kinases controlling the activity of the response regulator LuxO through phosphorylation (30, 35, 51). LuxO, in turn, activates the expression of four small RNAs (qrr1 to -4) that control the expression of the transcriptional regulator HapR (also called LuxR in V. harveyi and not related to the V. fischeri LuxR protein) by influencing the stability of its mRNA (29). At high cell density, LuxO is unphosphorylated and unable to activate the expression of the small RNAs, thus allowing HapR and LuxR to accumulate and influence gene expression.
V. cholerae HapR and V. harveyi LuxR proteins regulate a number of important cellular processes via quorum sensing. At high cell density, HapR represses both virulence gene expression (35, 51) and biofilm formation (16). This regulation is the reverse of most other quorum-sensing systems in which virulence and biofilm formation are induced at high cell density and is thought to contribute to the self-limiting nature of cholera infections. HapR represses virulence gene expression by binding to a specific site in the aphA promoter between positions −85 and −58 (25). This reduces the intracellular levels of AphA and prevents activation of the virulence cascade. The mechanism by which HapR represses biofilm formation is not yet known. HapR also activates the expression of a hemagglutinin referred to as HA/protease (20) and a newly discovered transcriptional regulator involved in natural competence for DNA uptake (32). LuxR regulates bioluminescence in V. harveyi by activating the expression of the luxCDABE operon (47).
V. cholerae HapR is a dimeric, 203-residue (23.6-kDa) protein that is 71% identical to V. harveyi LuxR (20). To further understand the molecular basis of HapR-mediated regulation of virulence gene expression, we have determined its crystal structure to 2.2-Å resolution. The all-helical structure reveals the presence of an N-terminal helix-turn-helix (HTH) DNA-binding motif and a large C-terminal dimerization domain that is similar to a number of other TetR family regulators (9, 17, 37, 43). The dimerization interface contains a unique solvent accessible tunnel that connects to an amphipathic cavity in each monomer and which may serve as a ligand-binding pocket for an unidentified small molecule effector.
MATERIALS AND METHODS
Protein preparation and crystallization.
The purification of HapR using the IMPACT-CN protein fusion and purification system (New England Biolabs) was previously described (31). The protein was concentrated to 5 mg ml−1 using Centricons (Millipore) and dialyzed overnight in 20 mM Tris-HCl (pH 7.6), 1 mM EDTA, 200 mM NaCl, and 0.1 mM dithiothreitol. HapR crystals were obtained using the vapor diffusion method by mixing equal volumes of the protein as described above with 0.2 M HEPES (pH 7.5), 12% (wt/vol) polyethylene glycol 8000, and 5% (vol/vol) ethylene glycol as the crystallization buffer. To determine the structure using the multi-wavelength anomalous dispersion method (MAD), a heavy atom derivative was obtained by soaking the native crystals in 1 mM ethylmercuricthiosalisylic acid (EMTS) for 6 h. For both native and derivative crystals, 20% glycerol was used as the cryoprotectant.
Data collection and structure solution.
A MAD data set was collected at NSLS beamline X6A at three wavelengths (peak, 1.00595 Å; inflection, 1.009068 Å; and remote, 0.93927 Å). After analysis with the program XDS (22), it was noted that the anomalous signal was absent in the second and third wavelength sweeps. Although unusual, such loss of anomalous signal is not unprecedented and has been attributed to radiation-induced cleavage of S-Hg bonds (41). Based on this phenomenon, a technique has been described for solving crystal structures using radiation damage-induced phasing with anomalous scattering (RIPAS) (41). We therefore adjusted our solution strategy to use this technique using the Hg-peak sweep as the “derivative” data set and the inflection sweep as the “native” data set (Table 1). Using these data, phases were obtained, the structure was solved, and an initial electron density map was calculated to 2.9 Å using the SOLVE/RESOLVE program (49) with scripts modified from a standard SIRAS (for single isomorphous replacement with anomalous scattering) solution. The quality of the resulting electron density map was excellent (Fig. 1), and the presence of a large number of tryptophan, tyrosine, and phenylalanine side chains allowed placement of the HapR sequence into the map. Approximately 70% of the model was built by using the software package O (21), and this starting model was completed by iterative rounds of refinement with CNS (3), followed by model rebuilding using 2Fo-Fc and Fo-Fc electron density maps. A 2.2-Å native data set (Table 1), collected at NSLS beamline X6A, was used to complete the model by iterations of manual rebuilding with the program COOT (11) and refinement using REFMAC5 (1) with four TLS groups made up of the two DNA-binding regions (A5 to A59, B5 to B59) and two dimerization domains (A60 to A199, B60 to B201). The final structure contains amino acids A5 to A201 in one monomer and B5 to B201 in the other monomer and has an Rwork value of 23.0% and an Rfree value of 26.3%. (see Table 1). A composite omit map, generated with CNS, shows the main chain density to be complete for the entire structure; however, side chain density is missing for residues A37, A123, A125, A156, A157, B37, B99, B123, and B185. Analysis of the final structure with PROCHECK showed 91.0% of residues in the most favored regions and 9.0% in additionally allowed regions. No residues were in generously allowed or disallowed regions.
TABLE 1.
Data collection, phasing, and refinement statistics
| Category | Data set
|
||
|---|---|---|---|
| Native used for refinement | “Native” used for phasing | “Derivative” used for phasing | |
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Unit cell dimensions (Å) | a = 43.8, b = 85.1, c = 110.4; α = β = γ = 90° | a = 44.8, b = 86.2, c = 110.2; α = β = γ = 90° | a = 44.8, b = 86.2, c = 110.2; α = β = γ = 90° |
| Resolution range (Å)a | 67.2-2.2 (2.3-2.2) | 20-2.9 (3.0-2.9) | 20-2.5 (2.6-2.5) |
| Wavelength (Å) | 1.0093 | 0.9392 | 1.0059 |
| Measured reflections | 86,355 (10,507) | 93,638 (9,120) | 144,148 (16,022) |
| Unique reflections | 42,360 (5,265) | 18,234 (1,783) | 28,427 (3,118) |
| Multiplicity | 2.0 (2.0) | 5.1 (5.1) | 5.1 (5.1) |
| Completeness (%) | 98.3 (98.3) | 99.6 (100.0) | 99.6 (100.0) |
| Rsymb (%) | 4.7 (30.9) | 5.0 (28.2) | 7.5 (24.8) |
| Phasing | |||
| Resolution range (Å) | 19.79-2.9 | ||
| Overall figure of merit | 0.70 | ||
| Refinement | |||
| Resolution range (Å) | 67.2-2.2 | ||
| Rcryst/Rfree (%) | 23.0/26.3 | ||
| RMS deviation bonds (Å)/angles (°) | 0.041/3.189 | ||
| No. of atoms (protein/solvent) | 3,237/322 | ||
| Avg B factors (Å2) | 47.3 | ||
Values in parentheses are for data in the highest-resolution shell.
Rsym = 100 × ΣhΣi | I(h) − Ii(h) | / ΣhΣi Ii(h), where Ii(h) and I(h) values are the ith and mean measurements of the intensity of reflection h.
FIG. 1.
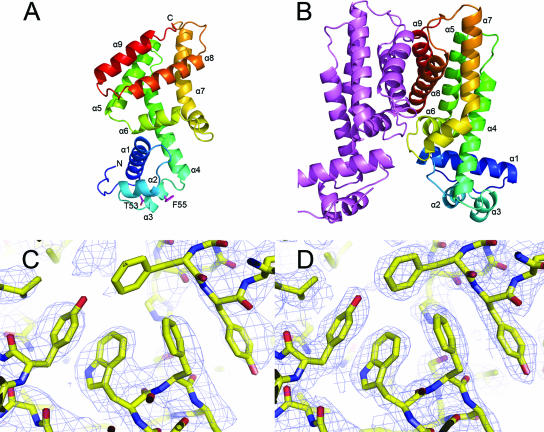
Structure of HapR. (A) HapR monomer viewed from the side. The DNA binding surface is at the bottom in this orientation. Helices are rainbow-colored according to the secondary structure succession from blue for the N-terminal helix α1 to red for C-terminal helix α9. Secondary structural elements are labeled accordingly. Helix α3 residues T53 and F55, which have been mutated in the present study, are shown in magenta. The N and C termini are indicated. (B) Side view of the HapR dimer, with one chain in magenta and one chain colored as in panel A. (C) View of the final HapR model showing 2.9-Å electron density at 1.0σ from the initial, experimentally phased, and solvent-flattened electron density map obtained directly from RESOLVE (49). (D) The same view of the final HapR model showing 2.2-Å electron density from a composite omit map calculated with CNS (3) using the final model, contoured at 1.5σ. The structural figures in the present study were rendered by using the program PYMOL (6).
Construction of point mutations and immunoblot analysis.
The point mutations in hapR were constructed by overlapping PCR using the primers H53C (5′-GATCGCTCTTCGGTGTTCAACTACTTCCCAACTC) and H55C (5′-GATCGCTCTTCGAACTACTTCCCAACTCGTGAAG) with HapX (5′-GATCGTCTAGACGGATGCGCCCTTTGTGCTG) and the primers H53A (5′-GATCGCTCTTCGCACTGCTGCAACGGAGACTTGCGCAATC) and H55A (5′-GATCGCTCTTCGGTTGGCCACTGTTGCAACGGAGACTTG) with HapE (5′-GATCGGAATTCACCATGGACGCATCAATCGAAAAACG). The resulting fragments were ligated into pMMB66EH (14), and the mutations were confirmed by DNA sequencing. The mutant hapR genes were then PCR amplified as previously described for the wild-type hapR gene (31) and ligated into pTXB1, generating pWEL121 (T53A) and pWEL122 (F55A). The mutant proteins were purified as described for the wild-type protein (31). The point mutations were also inserted into the vector pKAS154 (25), generating pWEL129 (T53A) and pWEL130 (F55A). They were then introduced into the chromosome of the V. cholerae aphA-lacZ fusion GK178 (25) by allelic exchange (46). Whole-cell extracts of HapR and its mutant derivatives in GK178 were prepared by growing the strains under AKI conditions (19) at 37°C for 5.5 h. After centrifugation and resuspension in sodium dodecyl sulfate sample buffer, they were subjected to sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis, transferred to nitrocellulose, and visualized by using an enhanced chemiluminescence detection system (Amersham). Anti-HapR antibody was generated at the Pocono Rabbit Farms from the purified protein (31) according to established protocols. The stability of the F55A mutant was also assessed by comparing the red shift in tryptophan fluorescence emission wavelength to wild-type protein in the presence of urea concentrations up to 7.2 M. The data (not shown) indicate similar stabilities for the wild-type and mutant proteins.
Gel mobility shift and β-galactosidase assays.
The 117-bp DNA fragment for the gel mobility shift assays was amplified by PCR from the aphA promoter of V. cholerae C6706 str2 using primers extending from −173 (YF13) to −56 (YF22) relative to the transcriptional start (25). The fragment was gel purified and end labeled with digoxigenin as previously described (24). Binding reactions for HapR were as previously described (25). The samples were applied to a 5% polyacrylamide gel and subjected to electrophoresis in 0.5× Tris-borate-EDTA at room temperature. The DNA was transferred to nylon membranes by electroblotting, probed with anti-digoxigenin-AP antibody (Amersham Pharmacia), and visualized by using chemiluminescence. β-Galactosidase assays were carried out as previously described (33).
The atomic coordinates and structure factors (PDB #2PBX) for HapR have been deposited in the Protein Data Bank (PDB), Research Collaboratory for Structural Bioinformatics (http://www.rscb.org).
RESULTS AND DISCUSSION
Overall structure and similarity to other TetR regulators.
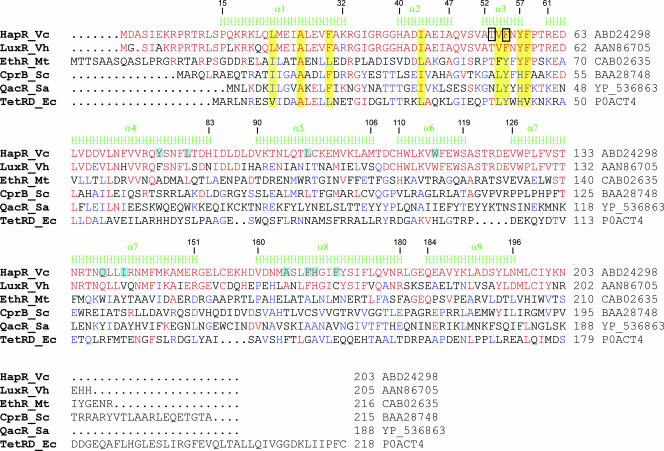
The crystal structure of HapR was determined by RIPAS (41) and subsequently refined to 2.2-Å resolution (Table 1 and Fig. 1A and B). The asymmetric unit contains a dimer in which the two subunits are related by a noncrystallographic twofold rotation axis. Each monomer of HapR is 31 Å long by 28 Å wide by 56 Å deep, resulting in a dimer with overall dimensions of 64 Å by 28 Å by 56 Å. Each monomer contains nine α helices: α1 (15 to 32), α2 (40 to 47), α3 (52 to 57), α4 (61 to 83), α5 (90 to 106), α6 (110 to 119), α7 (126 to 151), α8 (160-180), and α9 (184 to 196) (Fig. 1A and B). The first three helices of each HapR monomer form a putative DNA-binding domain, containing a characteristic HTH motif (α2 and α3). The large C-terminal domain of HapR is composed of six α helices (α4 to α9) and is involved in the dimerization of the repressor. The overall fold of HapR is very similar to other members of the TetR superfamily, including the Escherichia coli tetracycline repressor TetR (17, 38), Staphylococcus aureus multidrug binding transcriptional regulator QacR (43, 44), Streptomyces coelicolor γ-butyrolactone autoregulator receptor protein CprB (36, 37), and Mycobacterium tuberculosis ethA repressor EthR (9, 13). These proteins share the greatest degree of sequence similarity in the N-terminal DNA-binding domain (Fig. 2). The sequences of the C-terminal regulatory domains show low similarity despite their overall conserved three-dimensional fold, most likely reflecting differences in the specific ligands recognized by the proteins.
FIG. 2.
Multiple sequence alignment of TetR family members and secondary structure assignment for HapR based on its crystal structure. The HapR secondary structure is shown above the sequence. Residue numbering is to the right for all proteins and also above the secondary structure elements for HapR. The sequences were aligned by using CLUSTAL W (4) and formatted using ESPript (15). Residues identical to HapR are red, and residues identical in two or more family members are blue. The black boxes show the residues altered in the present study to alanine. Conserved residues in the DNA-binding domain discussed in the text are highlighted in yellow, and residues in the putative binding pocket (shown in Fig. 3C) are in highlighted in cyan. (Accession numbers are shown to the right of each line.) Vc, V. cholerae; Vh, V. harveyi; Mt, M. tuberculosis; Sc, S. coelicolor; Sa, S. aureus; Ec, E. coli.
In order to identify the closest structural homologues to HapR, database searches were performed using both DALI (18) and SSM (27). The top hits from the DALI search included seven uncharacterized, putative transcription factors whose structures have been determined by various structural genomics initiatives (PDB codes 2HYJ, 2G3B, 1VI0, 1ZK8, 1SGM, 1Z77, and 2GFN; DALI aligned HapR chain A with chain A of each of these). The eighth hit was a characterized protein, the multidrug binding transcriptional regulator QacR (42, 43). The SSM search also identified QacR as the most homologous protein for which a thorough characterization has been reported. Among the top SSM hits, the highest homology was between chain B of the HapR structure and the drug-free chains from a number of dimeric structures of QacR bound with cationic, lipophilic drugs, including ethidium (PDB code 1JTY, chain E), malachite green (1JUP:E), hexamidine (1RPW:D), berberine (1JUM:E), proflavine (1QVT:E), and dequalinium (1JT6:E). Also high on the SSM list was the drug-free, DNA-bound form of QacR (1JT0:B). For all of these structures, SSM reported Q scores between 0.50 and 0.46, with root mean square deviation values of 2.17 Å to 2.40 Å for between 161 and 171 aligned alpha carbons.
Dimerization domain and interface.
The dimerization interface is one of the conserved structural features of the TetR family and, as shown in Fig. 1B, in HapR forms by coupling α8 and α9 in one monomer to form a four-helix bundle with the same helices of the other monomer, burying over 1,700 Å2. Helices α8 and α9, which are 21 and 13 amino acids long and pack in parallel to each other, are stabilized by many hydrophobic contacts involving Tyr194, Leu166, Leu154, Leu176, Ala120, Tyr172, Trp127, and Leu181. Additional amino acid contacts are provided by the loops connecting helices α1 and α2, helices α6 and α7, and helices α7 and α8. Notable among these interactions, residues Arg37, Thr122, Arg123, and His158 form both hydrophobic and polar contacts. Tyr172 in helix α8 forms stacking interactions, and Cys199 in helix α9 forms a disulfide bridge with its complementary residue. Although it is unlikely that this disulfide bond is formed in the reducing environment of the cell interior, a similar disulfide bridge is present in the CprB structure via Cys159 on helix α8 between the two subunits (36). Other interesting interactions include a water-mediated hydrogen bonding network between Asp159 amide nitrogen of one monomer and Ser193 of the other monomer and a salt bridge between Glu117 of one monomer and Arg123 of the other monomer. The interactions between these parallel helices are duplicated by the twofold dimer symmetry, contributing to the stability of the HapR dimer.
Putative ligand-binding domain.
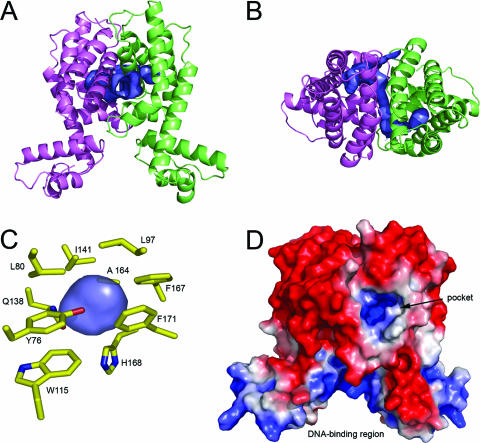
Several programs were used to identify cavities and putative ligand-binding sites in the HapR dimer. The program SURFNET (28) was used to visualize cavities and showed a tunnel ∼3.0 Å in diameter leading through the dimerization interface (Fig. 3A and B). Furthermore, in each monomer an amphipathic pocket is connected to this tunnel. The pockets are essentially identical in each monomer; however, in monomer A the pocket is solvent accessible, whereas in monomer B rotation of the side chain of Gln138 partially blocks the opening between the tunnel and the pocket. Analysis of the structure with the online program CASTp (10) shows the Connolly/molecular surface-based volume of the tunnel and monomer A pocket combined to be 1412 Å3 and the volume of the isolated monomer B pocket to be 130 Å3. In both monomers, this putative ligand-binding domain is surrounded by polar amino acid side chains on one side of the pocket, with hydrophobic residues on the opposite side (Fig. 3C). Analysis of the overall surface electrostatics (Fig. 3D) shows both the tunnel and the pockets to have a net positive charge, indicating that the binding ligand is likely to be amphipathic and contain anionic moieties. This is in marked contrast to QacR, in which the ligand-binding pocket is able to accommodate a structurally diverse set of cationic lipophilic drugs whose binding is stabilized by glutamate side chains (44). It should be noted that whereas all TetR family members whose structures have been solved contain ligand-binding pockets in a homologous location, the presence of a solvent accessible tunnel is unique to HapR.
FIG. 3.
View of the solvent accessible tunnel and putative ligand-binding pocket of HapR. (A) Ribbon diagram of the HapR dimer, oriented as in Fig. 1B, showing the location of the tunnel and cavity in blue, monomer A in magenta, and monomer B in green. The surfaces in this figure were generated by using the program SURFNET (28) and rendered by using PYMOL (6). (B) Top view of the tunnel and cavities looking down the HapR dimer interface. (C) View of residues containing atoms surrounding the solvent isolated pocket in monomer B. (D) Electrostatic surface potential of the HapR dimer, oriented as in panel A. Blue and red indicate positive and negative charges, respectively. The black arrow points to the electropositive entrance to the solvent accessible tunnel and pocket, visible slightly to the right of the center of the dimer. The electrostatic surface was calculated by using the APBS (2) plug-in to PYMOL (6). Atomic charges and radii were calculated by using the AMBER force field option at the online PDB2PQR service (8).
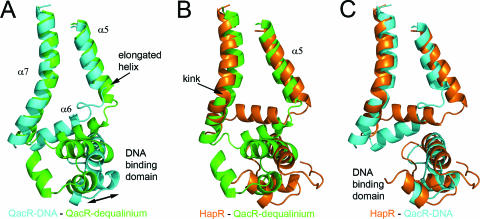
Since QacR showed the highest degree of structural similarity to HapR, we compared our structure to two forms of QacR: the drug-free, DNA-bound form, determined to 2.9-Å resolution (43), and the dequalinium-bound form, determined to 2.54-Å resolution (44). These two structures have been used to provide a model as to how drug binding in QacR induces structural changes resulting in its release from DNA (42). In the drug-free structure of QacR bound to DNA, a binding pocket of ∼400 Å3 is flanked by helices α5 and α6 (Fig. 4A). Binding of a drug, such as dequalinium, induces a coil-to-helix transition that lengthens helix α5 by one complete turn. This has two primary results: two tyrosine residues, acting as “drug surrogates,” are removed from the drug binding pocket, thereby increasing its volume to ∼1,100 Å3, and helix α6 moves, resulting in a rotation and translation of the DNA-binding domain. Comparison of HapR with these two conformations shows some interesting similarities, as well as some differences. Despite HapR being in a ligand-free state, helix α5 is extended, as in the drug-bound form of QacR (Fig. 4B). However, the relative position of the HapR DNA-binding domain is similar to that of the DNA-bound form of QacR (Fig. 4C), and we predict that the HapR dimer is in a conformation that would support DNA binding without significant conformational changes (see below).
FIG. 4.
Comparison of HapR with DNA-bound apo QacR and dequalinium-bound QacR. (A) SSM (27) produced superposition of DNA-bound apo QacR (PDB 1JT0, chain B) shown in cyan and dequalinium-bound QacR (1JT6:A) shown in green. For clarity, only helices α5, α6, and α7 (amino acids 76 to 136) and the DNA-binding domain (amino acids 2 to 4) are shown. Note that in the dequalinium-bound QacR structure, helix α5 is extended by a full turn, causing the DNA-binding domain to move in relation to helices α5 to α7, as indicated by the arrow. (B) Alignment of HapR, shown in orange, and QacR-dequalinium, shown in green, showing that helix α5 is extended in both of these structures to a similar degree and that the DNA-binding domains do not align. The pronounced kink in helix α7 of HapR is indicated. C, Alignment of HapR, in orange, and apo QacR-DNA, in cyan, showing the close alignment of the DNA-binding domains.
Although at first it appears that the structural logic described for QacR has not been followed by HapR, the presence of a kink in helix α7 provides an explanation for HapR having an extended α5, as well as a binding-competent position of its DNA-binding domain. The kink, which is significantly more pronounced than the bend observed in QacR structures (Fig. 1A and B and Fig. 4B), allows helix α6 to remain in an “up” position despite the presence of an extended helix α5. This suggests a model in which drug binding to HapR induces release from DNA via a straightening of helix α7, rather than the coil-to-helix transition in helix α5 observed in QacR. Loss of the kink would produce a movement of α6 and the DNA-binding domain into a position similar to that seen in drug-bound QacR, resulting in a structure that could no longer bind to DNA. While the absence in helix α5 of hydrophobic residues that could serve as “drug surrogates” also suggests that HapR operates under a different specific mechanism from QacR, it is interesting that the global changes are likely to be similar for both proteins as they switch between conformations that are able or unable to bind to DNA.
DNA-binding domain and model for HapR-DNA interaction.
The DNA-binding domain of HapR is composed of three α-helices, α1, α2 and α3, where helices α2 and α3 form a typical HTH motif. The residues that form the hydrophobic core of the DNA-binding domain (Leu22, Ala26, Phe30, I43, V54, and Phe58) are fairly well conserved among family members (Fig. 2). In both TetR (38) and QacR (43), helix α3 serves as the recognition helix that interacts with the major groove of target DNA. Similar to other family members, the C-terminal end of helix α3 in HapR is rich in aromatic residues (Phe55, Tyr57, and Phe58) (Fig. 2). Also, HapR contains a positively charged Arg61 as the first residue in helix α4, a position in which many other TetR family members have a conserved lysine that has been shown to contact the phosphate backbone in both TetR-DNA and QacR-DNA complexes (9).
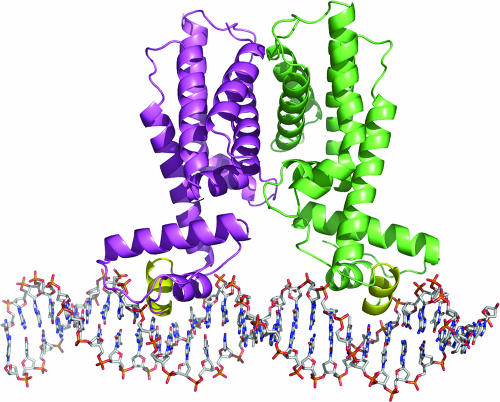
To predict what interactions HapR is likely to have with DNA, a model of the HapR-DNA complex was generated (Fig. 5) by aligning the HapR dimer with the QacR dimer-DNA structure (PDB code 1JT0, chains B and D) (43). Superposition of the two dimers results in a very close alignment, with 284 Cα atoms having an root mean square deviation of 2.3 Å. The alignment places the DNA-binding helices (α2 and α3) of HapR within 0.8 Å of the homologous helices in QacR, with the two DNA-binding domains being only 1.5 Å farther apart in the HapR dimer than in QacR. As expected, the DNA-binding surface of HapR displays a dominantly electropositive surface (Fig. 3D). The model predicts that residues in or near helix α3, including Ser50, Val51, Ala52, Thr53, Phe55, and Asn56, will form polar and/or hydrophobic contacts with the major groove DNA bases, whereas residues Arg10, Arg12, Arg18, Lys19, Arg33, Arg37, His40, and Arg61 are predicted to interact with the phosphate backbone.
FIG. 5.
Model of the HapR-DNA complex. A side view of the model of the HapR dimer (one chain in cyan and the other in green) bound to DNA is shown. The primary DNA interactions are predicted to be via helix α3 (yellow), which fit into adjacent major grooves on the same face of the DNA. The model was made by aligning the HapR dimer with the QacR dimer-DNA structure (PDB code 1JT0, chains B and D).
Mutational analysis of residues in helix α3.
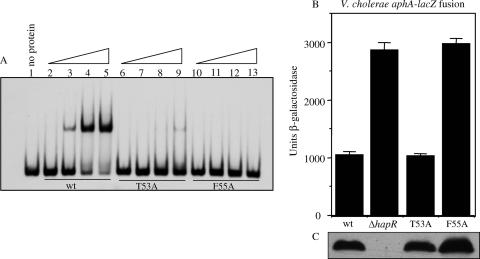
In order to further characterize the structural interactions governing DNA-binding affinity, two residues in helix α3 predicted to form contacts with DNA, Thr53 and Phe55, were altered to Ala. The mutant proteins were purified by using the same method that was used for the wild-type protein (31), and their ability to bind to DNA was assessed by using a gel mobility shift assay with a fragment of the aphA promoter that encompasses the HapR binding site. As shown in Fig. 6A, the wild-type protein bound strongly to this fragment but, at the same concentrations of protein, the F55A mutant was completely defective for binding, and the T53A mutant bound only very weakly. These results indicate that both of these positions are important for high-affinity binding of HapR to the aphA promoter.
FIG. 6.
Mutational analysis of residues in HapR helix α3. (A) Binding of wild-type and mutant HapR proteins to the aphA promoter. Lane 1, no protein added; lanes 2 to 5, wild-type HapR; lanes 6 to 9, HapR T53A; lanes 10 to 13, HapR F55A. The first lane in each set has 5 nM (2.5 ng) protein, the second lane has 50 nM (25 ng), the third lane has 200 nM (100 ng), and the fourth lane has 500 nM (250 ng). (B) Influence of HapR mutations on the expression of an aphA-lacZ promoter fusion in V. cholerae. Strains were grown under AKI conditions (19) at 37°C for 3.5 h. From left to right are shown GK178 (wt), KSK1815 (ΔhapR), WL521 (T53A), and WL522 (F55A). (C) Western analysis of crude extracts (17.5 μg) of the strains in panel B using anti-HapR antibody.
To determine whether the mutations also influenced the ability of HapR to repress virulence gene expression and biofilm formation in vivo, both mutations were introduced into the chromosome of a V. cholerae aphA-lacZ promoter fusion strain. Surprisingly, the T53A mutant retained the ability to repress the expression of the aphA promoter similar to that of wild-type HapR (Fig. 6B), whereas the F55A mutant was no longer capable of repressing its expression and appeared similar to that of a ΔhapR mutant. Consistent with these results, the F55A mutant exhibited a rugose colony morphology indicative of the overexpression of the Vibrio polysaccharide genes critical for biofilm formation that occurs when HapR is unable to repress their expression (16). In contrast, the T53A mutant showed a smooth colony morphology similar to the wild type. These results indicate that Phe55 is a critical residue for HapR binding to both the aphA and the vps promoters in vivo to repress virulence and biofilm formation, respectively. It may make a base-specific contact with DNA similar to that of the analogous residue in QacR, Tyr40 (43). Another example of a specific contact between an aromatic side chain and a nucleotide base has been observed in the crystal structure of a mutant form of the E. coli catabolite activator protein CAP-DNA complex that shows a phenylalanine can effectively replace a glutamate (E181) by forming a base-specific, aromatic hydrogen bond (39). Although the T53A mutation showed a defect in the more stringent in vitro binding assay, the mutant retained the ability to repress the expression of the aphA and vps promoters in vivo. Thus, this residue does not appear to be critical for the DNA-binding activity of HapR in vivo.
Concluding remarks.
Although HapR is not related to the TraR-type of quorum-sensing regulator that responds directly to autoinducer, the structure presented in the present study reveals that HapR possesses a unique solvent accessible tunnel that may accommodate an as-yet-unidentified small molecule ligand. This raises the intriguing possibility that, in addition to its regulation by quorum sensing, which influences its intracellular levels, a second level of regulation may influence its DNA-binding activity. For example, at high cell density, when HapR levels are increased, the binding of a particular ligand to HapR may induce conformational changes in the protein similar to those observed in other TetR regulators, which releases the protein from the DNA, thereby inducing the expression of virulence and biofilm genes. Studies to identify a ligand are currently in progress.
Acknowledgments
This study was supported by National Institutes of Health grants AI060031 and AI072661 (to F.J.K.), AI39654 (to R.K.T.), and AI41558 (to K.S.).
We thank the staff of NSLS beamline X6A, particularly Vivian Stojanoff, for help with data collection and Jean Jakoncic for help in analyzing the RIPAS data set. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract DE-AC02-98CH10886.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Anonymous. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 2.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 4.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 6.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 7.De Silva, R. S., G. Kovacikova, W. Lin, R. K. Taylor, K. Skorupski, and F. J. Kull. 2005. Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily. J. Biol. Chem. 280:13779-13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolinsky, T. J., J. E. Nielsen, J. A. McCammon, and N. A. Baker. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32:W665-W667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dover, L. G., P. E. Corsino, I. R. Daniels, S. L. Cocklin, V. Tatituri, G. S. Besra, and K. Futterer. 2004. Crystal structure of the TetR/CamR family repressor Mycobacterium tuberculosis EthR implicated in ethionamide resistance. J. Mol. Biol. 340:1095-1105. [DOI] [PubMed] [Google Scholar]
- 10.Dundas, J., Z. Ouyang, J. Tseng, A. Binkowski, Y. Turpaz, and J. Liang. 2006. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34:W116-W118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 12.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 81:4154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frenois, F., J. Engohang-Ndong, C. Locht, A. R. Baulard, and V. Villeret. 2004. Structure of EthR in a ligand bound conformation reveals therapeutic perspectives against tuberculosis. Mol. Cell 16:301-307. [DOI] [PubMed] [Google Scholar]
- 14.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 15.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 16.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 17.Hinrichs, W., C. Kisker, M. Duvel, A. Muller, K. Tovar, W. Hillen, and W. Saenger. 1994. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science 264:418-420. [DOI] [PubMed] [Google Scholar]
- 18.Holm, L., and C. Sander. 1996. Mapping the protein universe. Science 273:595-603. [DOI] [PubMed] [Google Scholar]
- 19.Iwanaga, M., K. Yamamoto, N. Higa, Y. Ichinose, N. Nakasone, and M. Tanabe. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30:1075-1083. [DOI] [PubMed] [Google Scholar]
- 20.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 21.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt. 2):110-119. [DOI] [PubMed] [Google Scholar]
- 22.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. [Google Scholar]
- 23.Kovacikova, G., W. Lin, and K. Skorupski. 2004. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol. Microbiol. 53:129-142. [DOI] [PubMed] [Google Scholar]
- 24.Kovacikova, G., and K. Skorupski. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB, and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41:393-407. [DOI] [PubMed] [Google Scholar]
- 25.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 26.Kovacikova, G., and K. Skorupski. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 181:4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krissinel, E., and K. Henrick. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60:2256-2268. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski, R. A. 1995. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J. Mol. Graph 13:307-330. [DOI] [PubMed] [Google Scholar]
- 29.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 30.Lilley, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36:940-954. [DOI] [PubMed] [Google Scholar]
- 31.Lin, W., G. Kovacikova, and K. Skorupski. 2005. Requirements for Vibrio cholerae HapR binding and transcriptional repression at the hapR promoter are distinct from those at the aphA promoter. J. Bacteriol. 187:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meibom, K. L., M. Blokesch, N. A. Dolganov, C. Y. Wu, and G. K. Schoolnik. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824-1827. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 35.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 36.Natsume, R., Y. Ohnishi, T. Senda, and S. Horinouchi. 2004. Crystal structure of a gamma-butyrolactone autoregulator receptor protein in Streptomyces coelicolor A3(2). J. Mol. Biol. 336:409-419. [DOI] [PubMed] [Google Scholar]
- 37.Natsume, R., R. Takeshita, M. Sugiyama, Y. Ohnishi, T. Senda, and S. Horinouchi. 2003. Crystallization of CprB, an autoregulator-receptor protein from Streptomyces coelicolor A3(2). Acta Crystallogr. D Biol. Crystallogr. 59:2313-2315. [DOI] [PubMed] [Google Scholar]
- 38.Orth, P., D. Schnappinger, W. Hillen, W. Saenger, and W. Hinrichs. 2000. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 7:215-219. [DOI] [PubMed] [Google Scholar]
- 39.Parkinson, G., A. Gunasekera, J. Vojtechovsky, X. Zhang, T. A. Kunkel, H. Berman, and R. H. Ebright. 1996. Aromatic hydrogen bond in sequence-specific protein DNA recognition. Nat. Struct. Biol. 3:837-841. [DOI] [PubMed] [Google Scholar]
- 40.Piper, K. R., S. Beck von Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448-450. [DOI] [PubMed] [Google Scholar]
- 41.Ramagopal, U. A., Z. Dauter, R. Thirumuruhan, E. Fedorov, and S. C. Almo. 2005. Radiation-induced site-specific damage of mercury derivatives: phasing and implications. Acta Crystallogr. D Biol. Crystallogr. 61:1289-1298. [DOI] [PubMed] [Google Scholar]
- 42.Schumacher, M. A., and R. G. Brennan. 2002. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 45:885-893. [DOI] [PubMed] [Google Scholar]
- 43.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 21:1210-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 294:2158-2163. [DOI] [PubMed] [Google Scholar]
- 45.Skorupski, K., and R. K. Taylor. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31:763-771. [DOI] [PubMed] [Google Scholar]
- 46.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 47.Swartzman, E., M. Silverman, and E. A. Meighen. 1992. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J. Bacteriol. 174:7490-7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terwilliger, T. C. 2002. Automated structure solution, density modification and model building. Acta Crystallogr. D Biol. Crystallogr. 58:1937-1940. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, L., P. J. Murphy, A. Kerr, and M. E. Tate. 1993. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature 362:446-448. [DOI] [PubMed] [Google Scholar]
- 51.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]