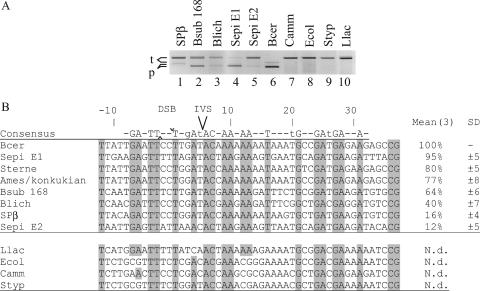
FIG. 5.
I-BanI can cleave several closely related Bacillaceae nrdE genes. (A) Agarose gel showing products (p) from a cleavage assay with fluorescein-labeled targets (t) from the B. subtilis 168 prophage SPβ (2,106 bp), B. subtilis 168 (2,103 bp), B. licheniformis (2,103 bp), S. epidermidis nrdE1 (2,103 bp) and nrdE2 (2,103 bp), B. cereus (1,174 bp), C. ammoniagenes (2,106 bp), E. coli (2,106 bp), S. enterica serovar Typhimurium (2,106 bp), and L. lactis (2,106 bp) incubated with I-BanI. Cleavage products corresponding to the predicted sizes were found for targets B. subtilis 168 prophage SPβ (1,073 bp), B. subtilis 168 (784 bp), B. licheniformis (784 bp), S. epidermidis nrdE1 (787 bp) and nrdE2 (766 bp), and B. cereus (722 bp) (lanes 1 to 6). No cleavage products were detected for the targets C. ammoniagenes, E. coli, S. enterica serovar Typhimurium, and L. lactis (lanes 7 to 10). Note that only one cleavage product was visualized, as only one strand was fluorescein labeled. (B) Target site alignments showing a region from positions −12 to +37 flanking the cuts on the coding strands of all target sequences tested. For each target, the measured relative activity is indicated, with the most efficiently cleaved target set as 100%. N.d. indicates no cleavage detected. Means and standard deviations (SD) were calculated from measurements from three experiments. DSB is indicated with ^ and ∨ for the cuts on the template and coding strands, respectively. Uppercase letters in the consensus sequence and gray boxes indicate fully conserved nucleotides within all cleaved targets. Lowercase letters in the consensus sequence indicate conserved nucleotides in target sequences cleaved 50% or more.