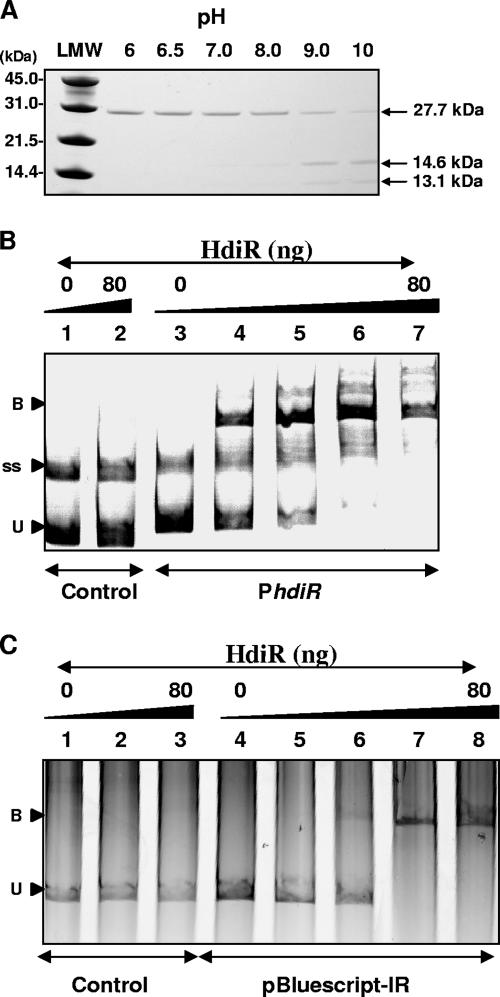
FIG. 7.
In vitro analyses of HdiR self-cleavage and DNA-binding activity. (A) pH-dependent cleavage of the His6-HdiR in the pH range of 6.0 to pH 10. Visualization of the autodigestion reactions containing 1,000 ng of the purified HdiR on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) gels stained with Coomassie brilliant blue. LMW refers to molecular weight standard (Bio-Rad). (B) Binding of the His6-HdiR to the putative promoter region of S. uberis hdiR-umuC. Reaction mixtures contained 35 ng of PCR-derived DNA fragments from the hdiR internal region (negative control, lanes 1 to 2) or from the upstream region of the hdir-umuC operon (lanes 3 to 7) mixed with 0 ng (lanes 1 and 3), 20 ng (lane 4), 40 ng (lane 5), 60 ng (lane 6), or 80 ng (lane 2 and 7) of HdiR. Reactions were separated by 5% PAGE followed by scanning of the gel with a Fujifilm FLA-5100 scanner using an excitation laser at 532 nm and an LPG emission filter. Positions of unbound (or free) DNA (U), protein-DNA complexes (B), and the single-stranded DNA probe (ss) are indicated. (C) Binding of the His6-HdiR to the pBluescript-II SK+ multiple cloning site containing a 26-bp control sequence without IR (lanes 1 to 3) or a 26-bp IR sequence (lanes 4 to 8) from the promoter region of hdiR. Reaction mixtures contained 35 ng of PCR-derived DNA fragments from pBluescript control (lanes 1 to 3) and from pBluescript-IR (lanes 4 to 8) mixed with 0 ng (lanes 1 and 4), 10 ng (lane 5), 20 ng (lane 6), 40 ng (lane 2), 60 ng (lane 7), and 80 ng (lanes 3 and 8) of HdiR. Reactions were separated in a 5% PAGE followed by staining with ethidium bromide and scanning with a Fuji FLA-5100 scanner using an excitation laser at 532 nm and an LPG emission filter. The positions of unbound (or free) DNA (U) and protein-DNA complexes (B) are indicated.