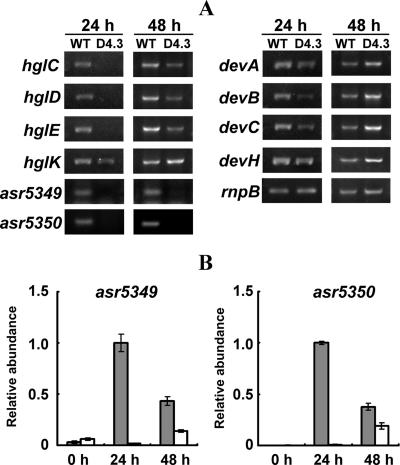
FIG. 6.
Analysis of transcripts of genes related to HGL synthesis or deposition. (A) Analyses by RT-PCR in the wild type (WT) and the mutant D4.3 strain. Total RNA was extracted from samples grown for 24 h and 48 h after deprivation of combined nitrogen. After cDNA synthesis, PCR was carried out for 20, 22, 24, and 26 cycles for rnpB; 24, 26, and 28 cycles for hglEACDK and devABC; 21, 23, and 25 cycles for devH; and 23, 25, and 27 cycles for asr5349 and asr5350. PCR products were loaded on a 1.2% agarose gel and separated by electrophoresis. For hglDEAK and devABC, the results are shown after 24 cycles; for rnpB, hglC, and devH, the results are shown after 22, 26, and 21 cycles, respectively; and for asr5349 and asr5350, results of PCR are shown after 27 cycles. The rnpB gene served as a control for the amount of RNA template. (B) Transcriptional analysis of asr5349 and asr5350 by real-time quantitative PCR in the wild type (gray bars) and D4.3 (open bars). The transcript levels were normalized to those of rnpB; the transcript level of wild-type cells at 24 h after the deprivation of combined nitrogen was set to 1, and other data were calculated according to this value.