Abstract
Magnetotactic bacteria navigate along the earth's magnetic field using chains of magnetosomes, which are intracellular organelles comprising membrane-enclosed magnetite crystals. The assembly of highly ordered magnetosome chains is under genetic control and involves several specific proteins. Based on genetic and cryo-electron tomography studies, a model was recently proposed in which the acidic MamJ magnetosome protein attaches magnetosome vesicles to the actin-like cytoskeletal filament formed by MamK, thereby preventing magnetosome chains from collapsing. However, the exact functions as well as the mode of interaction between MamK and MamJ are unknown. Here, we demonstrate that several functional MamJ variants from Magnetospirillum gryphiswaldense and other magnetotactic bacteria share an acidic and repetitive central domain, which displays an unusual intra- and interspecies sequence polymorphism, probably caused by homologous recombination between identical copies of Glu- and Pro-rich repeats. Surprisingly, mamJ mutant alleles in which the central domain was deleted retained their potential to restore chain formation in a ΔmamJ mutant, suggesting that the acidic domain is not essential for MamJ's function. Results of two-hybrid experiments indicate that MamJ physically interacts with MamK, and two distinct sequence regions within MamJ were shown to be involved in binding to MamK. Mutant variants of MamJ lacking either of the binding domains were unable to functionally complement the ΔmamJ mutant. In addition, two-hybrid experiments suggest both MamK-binding domains of MamJ confer oligomerization of MamJ. In summary, our data reveal domains required for the functions of the MamJ protein in chain assembly and maintenance and provide the first experimental indications for a direct interaction between MamJ and the cytoskeletal filament protein MamK.
For navigation along the earth's magnetic field, magnetotactic bacteria (MTB) use unique organelles, the magnetosomes, which in most MTB are membrane-enclosed crystals of the magnetic iron mineral magnetite (Fe3O4). Intracellular vesicles formed by the magnetosome membrane (MM), which are synthesized prior to magnetite formation by invagination from the cytoplasmic membrane (14, 26), represent a distinct subcellular compartment that provides spatial and physicochemical control over magnetite crystal formation. A specific subset of proteins was identified in the MM of Magnetospirillum gryphiswaldense and related MTB, which have presumed functions in vesicle maturation, magnetosome-directed iron transport, crystal formation, and magnetosome chain formation (6, 7, 30, 31).
Efficient geomagnetic field orientation of MTB depends on highly ordered linear chains of magnetosome particles, which transmit the rotational magnetic torque to the cell body (2). However, the assembly and stabilization of magnetosome chains have remained a puzzle, because magnetic dipoles tend to arrange into more energetically favored assemblages such as rings and aggregates, unless stabilized by a biological structure (11, 12, 21). Recently, several studies addressed the mechanism of magnetosome chain assembly at the molecular level. It was reported by Scheffel and coworkers that cells of Magnetospirillum gryphiswaldense, in which the gene encoding the acidic magnetosome protein MamJ was deleted, no longer assemble linear chains of magnetosomes, but instead magnetosomes are arranged in three-dimensional clusters (26). Cryo-electron tomography (cryo-ET) revealed that in M. gryphiswaldense wild-type cells, empty and mature magnetosomes were attached to a novel filamentous structure that extended from pole to pole adjacent to the cytoplasmic membrane. The observation that empty magnetosome vesicles of MamJ-deficient cells in contrast were detached from the cytoskeletal magnetosome filament and scattered throughout the cytoplasm (26) led to a model in which MamJ connects magnetosome vesicles to the magnetosome filament, possibly by interaction through its conspicuous CAR domain, thereby preventing the magnetosome chain from collapsing. Because of its striking sequence homology to bacterial actin-like proteins (31), it was further speculated by Scheffel et al. (26) that the magnetosome filament is formed by the product of the mamK gene, which is cotranscribed with the adjoining mamJ gene within the mamAB operon of the genomic magnetosome island (29, 37). The first experimental data supporting this hypothesis were provided by Komeili and coworkers (14), who generated a ΔmamK mutant strain of Magnetospirillum magneticum which they analyzed by cryo-ET. In tomograms of ΔmamK mutant cells, the magnetosome filament was not apparent, suggesting its absence to be connected to the deletion of the mamK gene (14). Recently, Pradel and coworkers (24) expressed green fluorescent protein (GFP)-tagged MamK of M. magneticum in Escherichia coli and observed the formation of a linear MamK-GFP filament which is structurally distinct from other prokaryotic actin-like filaments as formed by MreB or ParM. Taken together, the reported observations indicate MamK serves as a filamentous scaffold for stabilizing magnetosome chains in a manner as proposed by Scheffel and coworkers (26, 27), but experimental data confirming the suggested physical interaction between MamJ and MamK have not been provided yet.
In order to gain better understanding of the mechanisms directing magnetosome chain assembly and to address the role of the conspicuous acidic repetitive domain of MamJ, we analyzed the domain structure of the MamJ protein with respect to its function in chain formation in greater detail. Various truncated MamJ proteins were constructed, which enabled us to map essential regions of MamJ by assaying the potential of mutant proteins to restore magnetosome chain formation in the ΔmamJ strain. We further present the first evidence for a physical interaction between MamJ and MamK and show that C- and N-terminal sequence regions mediate interaction, whereas the CAR domain apparently is dispensable for MamJ function under all tested conditions.
MATERIALS AND METHODS
Bacterial strains and media.
Strains Magnetospirillum gryphiswaldense MSR-1 (DSM6361), strain MSR-1ΔmamJ (26), strain MSR-1B (28), Magnetospirillum magnetotacticum MS-1 (ATCC 31632), and the two novel isolates Magnetospirillum sp. strains CF-2 and CF-3, which are closely related to Magnetospirillum sp. strain MSM-6 (5, 32), were used. Liquid cultures of all Magnetospirillum strains were grown in modified FSM medium (8). For growth of Escherichia coli strain BW29427 (a dap auxotroph derivative of strain B2155 kindly provided by Barry Wanner), LB broth was supplemented with dl-α,ɛ-diaminopimelic acid (Sigma-Aldrich, Switzerland) to 1 mM. Medium for the E. coli BacterioMatch II validation reporter strain (Stratagene, La Jolla, CA) was prepared according to the instruction manual of the BacterioMatch II kit. Culture conditions for E. coli strains were as previously described (25).
DNA techniques.
Total DNA from all Magnetospirillum strains used in this study was isolated as described previously (19). Genetic constructs used in complementation and two-hybrid studies were generated using standard PCR procedures. The primers and plasmids used in this study are shown in Tables S1 and S2 in the supplemental material. Primer sequences for amplification of DNA fragments from M. gryphiswaldense MSR-1 were deduced from GenBank sequence deposition BX571797. The primer pair used for the amplification of mamJ from Magnetospirillum strains MS-1, CF-2, and CF-3 was deduced from sequence deposition NZ_AAAP01003824.1. For sequencing, BigDye terminators v3.1 (Applied Biosystems, Darmstadt, Germany) were used. Sequence data were analyzed with Lasergene 6 (DNAstar, Inc., Madison, WI) and MacVector 7.0 (Oxford Molecular, Ltd., Oxford, United Kingdom) programs.
Complementation studies.
For genetic complementation of the ΔmamJ mutant, full-length and mutated mamJ constructs were inserted in EcoRI-XbaI-restricted pBBR1MCS-2 (see Table S1 in the supplemental material) and introduced into the recipient strain M. gryphiswaldense MSR-1ΔmamJ by biparental conjugation with E. coli BW29427 as a donor. Kanamycin-resistant transconjugants of strain ΔmamJ were selected essentially as described previously (35). Full-length mamJ from M. gryphiswaldense was amplified using primer pair ASmamJs_f/ASmamJe_r1, and mamJ from strains MS-1, CF-2, and CF-3 was amplified using ASmamJ-MS1_f/ASmamJ-MS1_r. Genetic constructs encoding N-terminally truncated MSR-1 MamJ proteins JΔ1-24 and JΔ1-45 were amplified using primer pairs ASmamJ-N24AS_f/ASmamJe_r1 and ASmamJ-N45AS_f/ASmamJe_r1, respectively. Constructs encoding the C-terminally truncated MamJ proteins J1-392 and J1-386 were amplified with forward primer ASmamJs_f in combination with backward primers ASmamJ-34AS_r and ASmamJ-40AS_r, respectively. MamJ constructs containing internal deletions were generated by fusion PCR (9) of partially overlapping DNA fragments as a PCR template by using flanking primers ASmamJs_f/ASmamJe_r1. For construction of mamJ mutant construct JΔ81-256, fusion PCR was done with overlapping DNA fragments amplified by using primer pairs ASmamJs_f/ASmamJ-otr_r and ASmamJ-otr_f/ASmamJe_r1, for construct JΔ136-294 using ASmamJs_f/ASmamJ-oad_r and ASmamJ-oad_f/ASmamJe_r1, for construct JΔ293-334 using ASmamJs_f/ASmamJ-ala_r and ASmamJ-ala_f/ASmamJe_r1, and for JΔ335-360 using ASmamJs_f/ASmamJ-lag_r and ASmamJ-lag_f/ASmamJe_r1. Internal deletion constructs of MamJ have the following characteristics: JΔ81-256 lacks the tandem repeat consisting of two 88-amino-acid repeat units, JΔ136-294 lacks about 76% of the CAR domain, JΔ293-334 lacks the entire Ala-rich domain, and JΔ335-360 lacks the sequence region between the Ala-rich and Gly-rich domains. Cell growth and magnetic response (Cmag) of cultures were measured turbidimetrically at 565 nm as previously described (33). For practical purposes, Cmag = 0 was assumed for nonmagnetic cells. To inactivate cell motility, cell suspensions were either treated with formaldehyde (0.002 volume of a 37% aqueous formaldehyde solution [Fluka, Switzerland] was added) or heat inactivated (60°C for 20 min) prior to Cmag measurements. By transmission electron microscopy (TEM) inspection of more than 300 wild-type cells, we found that more than 99% of all cells had a linear, continuous chain of at least 10 magnetite crystals. Consequently, we considered complementation constructs capable of restoring the wild-type chain when ΔmamJ mutant cells containing magnetosomes were arranged in such a manner. Proper expression of the complementation constructs was verified by immunoblotting of protein crude extracts from complemented M. gryphiswaldense ΔmamJ cultures using a polyclonal antiserum raised against MamJ.
Construction of MamK-EGFP.
A mamK-egfp enhanced GFP (EGFP)-expressing fusion was generated by fusion PCR (9) using primer pairs ASmamKs_f/ASmamKe_r4 and ASegfp_f11/ASegfp_r3. The mamK-egfp fusion gene was cloned into the EcoRI and XbaI sites of vector pBBR1MCS-2 for expression. In the fusion protein, a -Gly-Ser- spacer lies between MamK and EGFP, the initiator ATG met codon of EGFP is replaced by the ATC ile codon, and the initiator CTG leu codon of MamK is replaced by the ATG met codon.
Immunoblot assay.
Western blotting was performed as previously described (34). For MamJ detection, a primary antibody was raised against the 15-amino-acid (aa) epitope APLAGNAESSEEGVV, which is present twice in full-length MamJ (one copy in each 88-aa repeat unit). Alkaline phosphatase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (CA).
Electron microscopy and fluorescence microscopy.
TEM was performed on a Zeiss EM 10 (PLANO, Wetzlar, Germany) with unstained cells adsorbed on carbon-coated copper grids. Fluorescence microscopy was performed on a Zeiss Axioplan 2 microscope equipped with a cooled charge-coupled device (CCD) camera. Images were acquired using Metamorph 6.3 (Universal Imaging Corp.) at exposure times of between 1 and 3.0 s. Image rescaling and cropping were done using Photoshop 6.0. Cell membranes were stained with the fluorescent dye FM4-64 (Molecular Probes) at a final concentration of 30 μM.
Bacterial two-hybrid assay.
Protein-protein interactions were investigated using the BacterioMatch II two-hybrid system vector kit and the BacterioMatch II validation reporter strain. To detect putative protein-protein interactions, the BacterioMatch II validation reporter strain was cotransformed with a pBT-derived bait expression vector (encodes fusion of bacteriophage λ repressor protein [λcI] with protein to test for protein binding activity) and a pTRG-derived prey expression vector (encodes fusion of the alpha subunit of E. coli RNA polymerase [αRNAP] with protein to test for protein binding activity). Growth of the cotransformants on selective screening medium plates containing 2.5 mM 3-amino-1,2,4-triazol (3-AT) was assessed according to the manufacturer's instructions. For construction of bait and prey expression vectors, full-length wild-type mamJ, full-length mamK, and partial mamJ sequences were in frame inserted into EcoRI-XhoI-restricted pBT and pTRG (see Table S1 in the supplemental material). Hence, expression vectors pBT_MamJ and pTRG_MamJ contained full-length wild-type mamJ, while pBT_MamK and pTRG_MamK harbored mamK. Bait and prey fusions with the N-terminal sequence of MamJ were encoded by vectors pBT_J1-135 and pTRG_J1-135. Only prey expression vectors were constructed with sequence regions encoding MamJ constructs J81-256 (pTRG_J81-256), J136-294 (pTRG_J136-294), J295-334 (pTRG_J295-334), J330-368 (pTRG_J330-368), and J361-399 (pTRG_J361-399). For construction of pTRG-based expression vectors for MamE (pTRG_MamE) and MamP (pTRG_MamE), mamE and mamP were inserted into EcoRI-XhoI sites of pTRG, while for MamA (pTRG_MamA) EcoRI-SpeI restriction sites of pTRG were used. Expression of pTRG encoding MamJ fusion constructs, containing the epitope targeted by the anti-MamJ peptide antibody, was confirmed by immunoblotting.
RESULTS
trans-complementation of the ΔmamJ mutant restores chain formation.
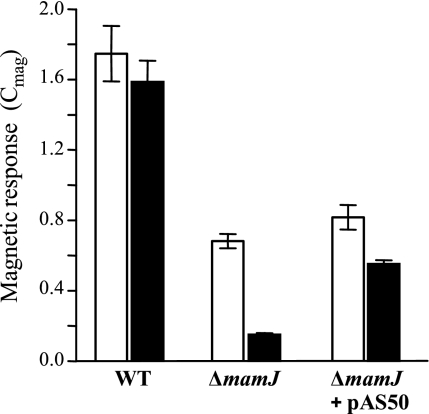
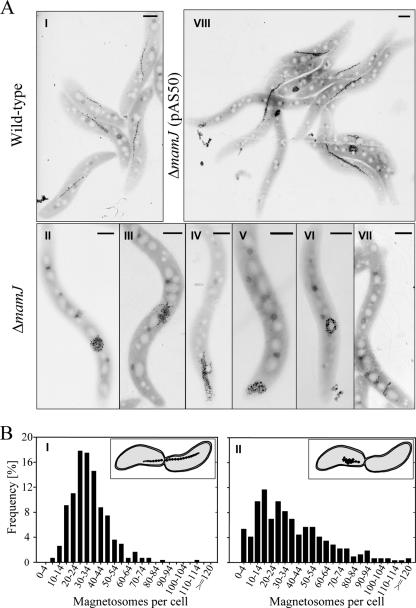
We previously noticed that a ΔmamJ mutant still aligns within magnetic fields, albeit magnetic response (Cmag) was reduced compared to that in the wild type (26). As Cmag measurements by means of light scattering are of critical importance for assaying the magnetic responses of different cultures, we systematically assessed this method for its applicability to quantify chain formation in ΔmamJ and trans-complemented ΔmamJ cells. To eliminate any effects of motility, cells were killed by the addition of formaldehyde or heat treatment. While Cmag values were comparable for viable and inactivated wild-type cells, the effect of formaldehyde killing was more pronounced in ΔmamJ and trans-complemented ΔmamJ cells (pAS50), suggesting that alignment along field lines is counteracted by active motility in those strains (Fig. 1). The same effect was observed when motility was inactivated by heat treatment of cells (data not shown). The magnetic response of formaldehyde-killed ΔmamJ cells (Cmag = 0.15 ± 0.01) was significantly reduced compared to that of wild-type cultures (Cmag = 1.59 ± 0.12), although they contained nearly identical average numbers of magnetosome particles (37 and 34, respectively). By inspection of 309 cells, we found the ΔmamJ strain to display a different distribution of magnetosome numbers per cell. In wild-type populations, magnetosome numbers exhibited a rather narrow symmetric distribution (μ = 34, σ = 14) (Fig. 2BI). Cells containing either very many (>60) or very few (<15) magnetosomes were only rarely observed and occurred at frequencies of only 4.21% and 3.34%, respectively. In contrast to the wild type, ΔmamJ cells exhibited a much wider distribution (Fig. 2BII), with a higher proportion of cells with very many (frequency, 17.24%) and very few or no (frequency, 19.12%) magnetosome crystals. ΔmamJ cells entirely devoid of magnetosome crystals occurred at a frequency of 3.5%. These divergent distributions of magnetosome numbers between wild-type and ΔmamJ cultures most likely result from asymmetric segregation of the magnetosome particles during cell division. A spot-like localization of magnetosomes as found in the magnetosome clusters of ΔmamJ cells more frequently will cause an “all-or-nothing” distribution of particles to the daughter cells than a linear chain, which is more likely to be divided evenly (Fig. 2BI and II, insets).
FIG. 1.
Cmag values (average magnetic response) of wild-type (WT), ΔmamJ, and ΔmamJ cells trans-complemented with the full-length wild-type allele (pAS50). Gray bars, motile cells; black bars, formaldehyde-killed cells. Cultures were grown in triplicate and diluted to an optical density at 565 nm of 0.1 prior to Cmag measurements.
FIG. 2.
(A) TEM micrographs of wild-type (I), ΔmamJ (II to VII), and ΔmamJ cells trans-complemented with pAS50 (VIII). Scale bars, 500 nm. (B) Distribution of magnetosome crystal numbers in stationary cultures of wild-type (I) and ΔmamJ (II) cells. Numbers were determined by counting 309 cells by TEM. Insets illustrate the magnetosome distribution to daughter cells during cell division in wild-type and ΔmamJ cells.
In addition to a larger proportion of cells containing no or only few magnetosomes, the reduced magnetic response of ΔmamJ cultures probably is also due to the nonlinear magnetosome arrangement, as irregularly arranged magnetosome particles partially or entirely will zero out their individual magnetic moments instead of adding up as in chains which occur in single, or occasionally, in duplicate in wild-type cells (Fig. 2AI). Besides clusters, we occasionally observed ring-like structures or imperfect short chains perpendicular to the cell long axis in ΔmamJ cells (Fig. 2AII to AVII), whose contribution to cellular magnetic response is adverse or uncertain.
In trans complementation of ΔmamJ cells with the full-length wild-type mamJ resulted in significant restoration of chain formation, as indicated by electron microscopy (Fig. 2AVIII). However, restoration of the wild-type phenotype was not complete, as only approximately 50% of the trans-complemented ΔmamJ cells produced wild-type-like chains, and Cmag values of trans-complemented cells consequently did not reach the wild-type level, which we attribute to artificial mamJ expression levels from a nonnative promoter or an effect of plasmid copy number.
Functional MamJ proteins display intra- and interspecies length polymorphism.
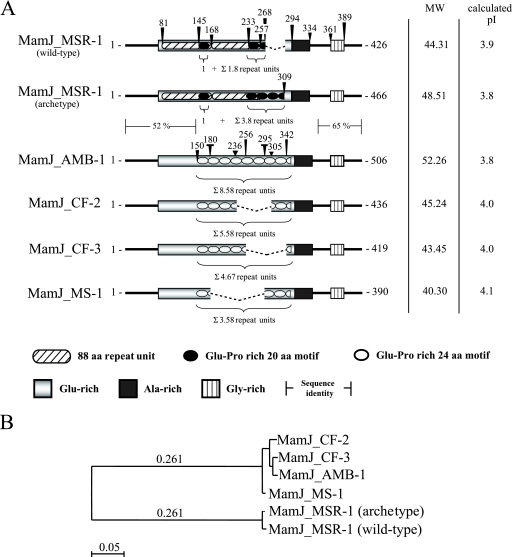
We noticed that PCR amplification of mamJ from various magnetic subcultures of Magnetospirillum gryphiswaldense MSR-1 yielded products of different lengths. For example, either a 1,281-bp product (in the following referred to as wild-type mamJ) or a 1,401-bp product (referred to as archetype mamJ) were amplified from the archetype and the lab strain, respectively. Sequencing of wild-type mamJ revealed an in-frame deletion encompassing a continuous 120-bp nucleotide stretch in the 3′-gene region. The absence of the 120-bp fragment apparently had no effect on chain formation, as cells harboring either archetype or wild-type alleles developed magnetosome chains that were virtually identical. The deduced amino acid sequence of both MSR-1 MamJ variants contains several distinct domains that display a highly biased amino acid composition (Fig. 3A). The most conspicuous sequence feature is the central acidic repetitive (CAR) domain, which comprises a direct repetition of an 88-aa motif (residues 81 to 168 and 169 to 256) followed by tandemly arranged copies of a highly acidic motif of 20 aa consisting primarily of Pro and Glu residues arranged in Glu-Pro segments. While the CAR domain of archetype MamJ comprises altogether 4.8 copies of the 20-aa motif (i.e., 2.8 copies located adjacent to the large repeat (residues 253 to 308) and one copy in each of the two 88-aa repeat units (residues 145 to 164 and 233 to 252), wild-type MamJ contains 2.8 copies because the deletion comprises a tandem copy of two 20-aa motifs (wild-type MamJ lacks residues 257 to 296 of archetype MamJ). In both MamJ variants, the CAR domain is followed by an Ala-rich domain and a Gly-rich domain is positioned near the C terminus, which is predicted to encompass a transmembrane segment (SAPS [http://bioweb.pasteur.fr/seqanal/interfaces/saps-simple.html], TopPred, TMpred, and TMAP [http://ca.expasy.org/tools/]).
FIG. 3.
Domain organization, sequence characteristics (A), and similarity tree (B) of MamJ protein sequences from M. gryphiswaldense MSR-1, M. magneticum AMB-1, M. magnetotacticum MS-1 and Magnetospirillum sp. strains CF-2 and CF-3. MW, molecular weight (in thousands).
To elucidate whether sequence variability within the CAR domain of MamJ is a general phenomenon, we compared MamJ sequences of M. gryphiswaldense MSR-1, M. magneticum AMB-1, M. magnetotacticum MS-1 and the two novel environmental isolates, Magnetospirillum strains CF-2 and CF-3 (5). In general, the sequence organization was found conserved in all homologs. However, sequence alignment showed MamJ of MSR-1 to be most divergent. While MamJ homologs from Magnetospirillum strains AMB-1, MS-1, CF-2, and CF-3 share more than 97% identity, sequence conservation to MamJ from MSR-1 is restricted to the first N-terminal 135 aa and the last C-terminal 76 aa, with 52% and 65% identity, respectively (Fig. 3B). Similar to MamJ variants of MSR-1, all other MamJ homologs display extensive tandem repeat polymorphism within the CAR domain. However, in MamJ from AMB-1, MS-1, CF-2, and CF-3, the tandem repeat consists of 24-aa units rich in Glu and Pro residues, which are repeated 8.6-, 3.6-, 5.6-, and 4.7 -fold, respectively. Despite sequence variations, all tested Magnetospirillum species displayed fully developed magnetic chains, and in trans expression of polymorphic mamJ homologs restored the formation of wild-type-like magnetosome chains in ΔmamJ cells, indicating that polymorphism of the CAR domain does not affect protein functionality.
The hypervariable CAR domain of MamJ is not required for magnetosome chain restoration in ΔmamJ cells.
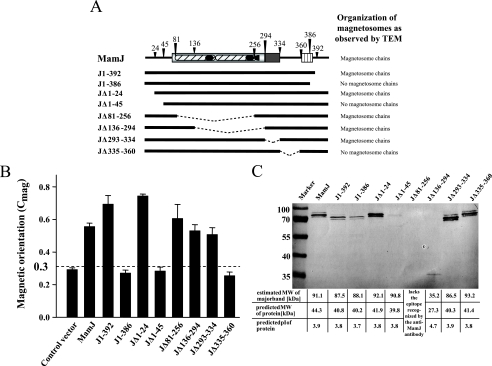
As the tandem repeat polymorphism within the CAR domain of MamJ has no obvious effect on the function of the protein, we were interested whether the hypervariable domain is required for magnetosome chain restoration at all. Therefore, we constructed various mamJ variants containing different deletions. To ensure expression of the various mamJ constructs, crude protein extracts of trans-complemented ΔmamJ strains were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). An antibody directed against MamJ recognized a band corresponding to 91.1 kDa, which is 2.06-fold greater than the calculated molecular mass of 44.3 kDa (Fig. 4C). Likewise, mutated MamJ proteins displayed aberrant migration behavior as well: C-terminally truncated proteins J1-392 and J1-386 migrated at 87.5 and 88.1 kDa, respectively; N-terminally truncated proteins JΔ1-24 and JΔ1-45 migrated at 92.1 and 90.8 kDa, respectively; and internal deletion-containing mutant proteins JΔ293-334 and JΔ335-360 migrated at 86.5 and 93.2 kDa, respectively (Fig. 4C). A strong shift in migration behavior (apparent molecular mass, 35.2 kDa) was observed upon deletion of about 76% of the CAR domain. For wild-type MamJ, the discrepancy between expected and actual electrophoretic mobility persisted after treatment with endo- and exoglycosidases or various strong denaturating agents, such as 8 M urea or 6 M guanidinium HCl (data not shown). Therefore, it is very unlikely that aberrant electrophoretic mobility is caused by incomplete dissociation of oligomers, which was previously suggested by Grünberg and coworkers (6). Instead, the observed shifts in mobility most likely result from the unusually high abundance of charged residues within the CAR domain of MamJ.
FIG. 4.
Ability of various MamJ mutant proteins to restore magnetosome chain formation in ΔmamJ. (A) Overview of constructs used for trans-complementation. Results of chain formation analysis by TEM are indicated by “+” or “−.” Dashed lines indicate deleted sequence fragments. (B) Cmag values (average magnetic response) of formaldehyde-killed ΔmamJ cultures trans-complemented with different MamJ mutant constructs. The dashed line represents a threshold (Cmag = 0.3) that discriminates between functional and nonfunctional MamJ mutant constructs. (C) Immunodetection of wild-type and mutant MamJ proteins. Proteins were expressed in trans in the ΔmamJ mutant and immunodetected by an anti-MamJ peptide antibody in crude extracts resolved by SDS-PAGE. Mutant protein JΔ81-256 lacks the epitope recognized by the antibody. Predicted molecular masses (MW) and isoelectric points are indicated below.
In order to identify essential domains of MamJ, ΔmamJ cultures expressing the various mamJ constructs were analyzed for restoration of magnetosome chain formation by TEM and our Cmag assay (Fig. 4A and B). TEM revealed that restoration of the wild-type phenotype was not complete and varied between cultures expressing different constructs. However, consistent with the entire absence of wild-type-like straight magnetosome chains in ΔmamJ strains expressing J1-386, JΔ1-45, and JΔ335-360 in TEM, these cells exhibited a significantly lower Cmag than cultures expressing MamJ constructs J1-392, JΔ1-24, JΔ81-256, JΔ136-294, and JΔ293-334, where wild-type-like chain formation was found restored in a significant proportion of cells.
In conclusion, restoration of magnetosome chains by JΔ81-256 and JΔ136-294 indicates that the hypervariable CAR domain is not essential for magnetosome chain restoration in ΔmamJ. Likewise, the Ala-rich domain (JΔ293-334) and also the first N-terminal residues (JΔ1-24) are not required to restore chain formation. However, regions indispensable for MamJ of MSR-1 are located in the N-terminal (residues 25 to 80) and in the C-terminal (residues 335 to 392) regions of the protein.
MamJ interacts with itself and with MamK.
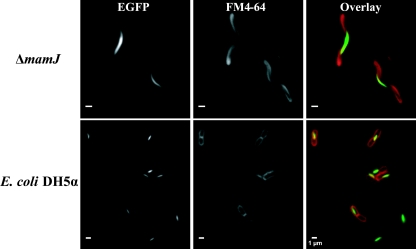
In the proposed model for magnetosome chain formation, MamJ at least transiently interacts directly with the filament-forming MamK protein (27). Since MamJ-EGFP does not properly localize in the 40-kb deletion mutant MSR-1B, lacking mamK (26), we asked whether the absence of MamJ on the other hand would affect the localization of MamK. However, our MamK-EGFP fusion displayed a localization pattern in the ΔmamJ mutant indistinguishable from that in the wild type (Fig. 5). Likewise, MamK-EGFP showed a linear localization pattern in Escherichia coli DH5α, indicating that filamentous localization of M. gryphiswaldense MamK does not depend on the presence of MamJ which has been previously reported by Pradel and coworkers (24). This finding is in accordance with the assumption that MamJ attaches magnetosomes to a MamK homofilament (27). In the wild type, the MamK-GFP fusion displayed localization virtually identical to that in the MSR-1B and ΔmamJ background (data not shown).
FIG. 5.
Intracellular localization of a MamK-EGFP fusion in ΔmamJ and in E. coli DH5α cells. Fluorescence micrographs of ΔmamJ cells and of DH5α cells stained with the membrane dye FM4-64 (red) and expressing a MamK-EGFP (green) fusion.
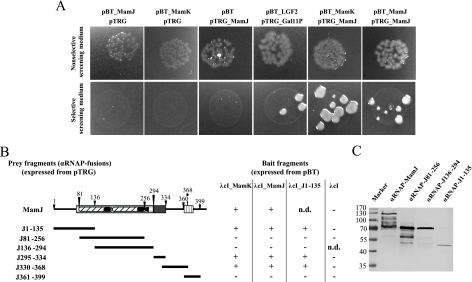
To provide experimental evidence for the proposed physical association between MamJ and MamK, we performed two-hybrid interaction analysis by means of commercial version of a prokaryotic two-hybrid system developed by Dove and coworkers (4). In the BacterioMatch II system, one protein of the pair of interest is expressed as C-terminal fusion with the λcI protein (bait fusion encoded on plasmid pBT), while the other is fused to the C terminus of αRNAP (prey fusion encoded on plasmid pTRG). Recombinant bait and prey expression vectors will be cotransformed into the E. coli reporter strain for expression of the protein fusions. In case both proteins interact, transcription of a chromosomally encoded reporter cassette consisting of a HIS3 reporter and an aadA gene is increased to levels sufficiently high to overcome competitive inhibition of the His3 enzyme by 3-AT, an additive of the selective screening medium, and AadA confers resistance to streptomycin as a secondary reporter.
Cotransformation of our bait (pBT_MamJ and pBT_MamK) and prey (pTRG_MamJ) expression vectors into the E. coli reporter strain yielded numerous colonies on nonselective screening medium, indicating that the transformation was successful (Fig. 6A). In contrast, cotransformants harboring pTRG_MamK only grew on nonselective screening medium when the isopropyl-β-d-thiogalactopyranoside (IPTG) concentration was reduced from 50 μM to 1 μM. This suggests that strong overexpression of the αRNAP-MamK fusion might be deleterious for the E. coli reporter strain. Fusion proteins λcI-MamJ and αRNAP-MamJ were found to interact because colony growth of cotransformed E. coli cells on selective screening medium was observed, which clearly indicates that MamJ does oligomerize (Fig. 6A). Colony formation on selective screening medium was also obtained for cells harboring the vector pair pBT_MamK/pTRG_MamJ, which indicates physically the interaction between the fusion proteins λcI-MamK and αRNAP-MamJ. To verify the observed positive interactions, we transferred patches of several colonies onto dual selective screening medium which contains 3-AT and streptomycin as a second inhibitor. We found transferred cells grew to colonies, which proves expression of the streptomycin resistance gene as consequence of a physical interaction between bait and prey fusion proteins. To ensure specificity of the observed interactions between αRNAP-MamJ/λcI-MamJ and αRNAP-MamJ/λcI-MamK, we further tested λcI-MamJ and λcI-MamK for interaction with αRNAP-MamA, αRNAP-MamE, αRNAP-MamP, αRNAP-Gal11P, and αRNAP. No colony growth on selective screening medium was found, which shows that neither λcI-MamJ nor λcI-MamK alone can self-activate expression of the reporter genes or physically interact with MamA, MamE, MamP, or Gal11P, respectively.
FIG. 6.
Two-hybrid analysis of the interaction between MamJ and MamK and MamJ and MamJ. (A) Growth of reporter E. coli spotted onto selective and nonselective screening medium after cotransformation with different bait (derivative of pBT) and prey (derivative of pTRG) expression vectors. Colony growth on nonselective screening medium verifies that cotransformation was successful, whereas on selective screening medium, colonies can grow only in the case of an interaction between bait and prey fusion proteins. (B) Overview of analyzed MamJ sequence regions (prey) for two-hybrid interaction with MamK, MamJ, and the N-terminal part of MamJ (bait). Growth on both selective and nonselective screening medium is shown by +; − indicates growth on nonselective screening medium only. n.d., not determined. (C) Immunodetection of prey fusion proteins by an anti-MamJ peptide antibody in protein crude extracts of the BacterioMatch reporter E. coli resolved by SDS-PAGE. RNAP fusions with J295-334, J330-368, and J361-399 lack the epitope recognized by the antibody.
C- and N-terminal sequence regions of MamJ are involved in oligomerization and binding to MamK.
Results of our complementation study described above already indicated that the hypervariable CAR domain of MamJ apparently is dispensable for maintaining protein functionality. To identify sequence regions of MamJ essential for either oligomerization or binding to MamK, we generated the prey expression vectors pTRG_J1-135, pTRG_J81-256, pTRG_J136-294, pTRG_J295-334, pTRG_J330-368, and pTRG_J361-399 by cloning various fragments of mamJ into plasmid pTRG. Cells of the E. coli reporter strain harboring either pBT_MamK or pBT_MamJ grew on selective agar plates when cotransformed with either pTRG_J1-135, pTRG_J295-334, or pTRG_J330-368 (Fig. 6B), indicating that protein binding activity of MamJ is inherent to its N-terminal sequence region and the sequence connecting the Gly-rich domain and the CAR domain. In contrast, the reporter strain did not form colonies on selective screening media after successful cotransformation of either pBT_MamK or pBT_MamJ in combination with either vector pTRG_J81_256, vector pTRG_J136-294, or vector pTRG_J361-399. In addition, no growth on selective medium was found after cotransformation of each prey expression vector together with pBT, indicating that fusions of αRNAP with MamJ fragments do not autoactivate transcription of the reporter cassette. In conclusion, these data provide additional evidence that neither the Glu-Pro-rich hypervariable CAR domain nor the C terminus of MamJ is involved in oligomerization or binding to MamK.
DISCUSSION
We further investigated the MamJ protein and its function in magnetosome chain assembly. A more comprehensive characterization of the ΔmamJ strain revealed that cells are not only impaired in chain formation but in addition display a different distribution of magnetic particles per cell compared to the wild type, which most likely results from asymmetric distribution of the aggregated particles during cell division. In addition to more-or-less-spherical clusters, we occasionally observed ring-like structures and imperfect chains. A ΔmamJ mutant could be functionally complemented by in trans expression of various homologous and heterologous MamJ variants as well as truncated MamJ fragments. Although trans-complementation did not fully restore the wild-type phenotype, the potential of chain formation could be monitored semiquantitatively by our light scattering assay. This was crucial for domain analysis by in trans-complementation, as allelic exchange of multiple fragments has remained a challenge due to poor efficiency of chromosomal insertions in magnetotactic bacteria.
The MamJ protein of Magnetospirillum species consists of three conserved domains of highly biased amino acid composition. The most prominent sequence feature of MamJ is the hypervariable CAR domain. We found extensive intra- and interspecies sequence variation within the CAR region between fully functional MamJ homologs of different Magnetospirillum species in the form of a tandem repeat polymorphism. For comparison, other magnetosome proteins such as MamK, MamA, or the CDF transporters MamB and MamM are much more conserved between various Magnetospirillum species, with identity levels of >93% (data not shown).
The tandem repeat polymorphism among MamJ homologs, which was probably caused by homologous recombination events between identical repeats, indicates that there is no or only low selective pressure for preservation of a defined structure to maintain protein functionality. On the other hand, variations in repeat copy numbers have been associated with variations in bacterial pathogenicity and human diseases and generate functional variability in yeast (20, 38). Thus, the observed variability might be a so-far-unrecognized source of genetic and phenotypic diversity of magnetosome organization and might provide a mean for rapid adaptation to the environment: e.g., by the modification of magnetosome organization as observed in other MTB species.
The conspicuous Glu-Pro-rich tandem repeat units within the CAR domain were expected to impart properties essential for the entire protein because clusters of charged residues increase protein solubility and contribute to both protein complex formation and repulsion between protein assemblages by electrostatic interactions (10), while on the other hand Pro-rich regions often are of structural importance (1, 39). Repetitions of Glu-Pro segments have been identified in a number of other eukaryotic and prokaryotic proteins. Although their exact function is mostly unknown, they are assumed to be of functional importance because of structural constraints that they confer upon the entire protein. For example, the bacterial periplasmic TonB protein, which is required for the uptake of several solutes, contains repeated Glu-Pro and Lys-Pro dipeptides separated by a short segment of 13 residues (3). Engineered TonB mutant proteins, in which the Pro-rich sequences were eliminated, retained functionality in E. coli cells grown under standard laboratory conditions. However, mutant TonB activity was reduced under osmolytic conditions that increase the periplasmic space (17, 18, 23), indicating that this domain is required for TonB function only under very specific environmental conditions. Contrary to our expectations, our data suggest no obvious function of this conspicuous sequence region, because engineered MamJ variants lacking these repeats were fully functional in restoration of magnetosome chain formation. However, we cannot exclude that the Glu-Pro-rich repeats confer a less obvious function that has not become apparent under the experimental conditions as described. For instance, nonnative protein levels due to in trans expression of MamJ might have compensated for more subtle phenotypes of MamJ alleles lacking the CAR domain, which could have escaped our notice. Thus, we cannot entirely rule out a possible regulatory function of the Glu-Pro-rich repeats, for instance, by modulating the strength of the vesicle attachment to the MamK-based filaments.
Two-hybrid systems have been widely applied not only to detect novel protein interaction partners but also to identify minimal domains or residues critical for interaction between defined protein pairs (16, 22). Despite the potential caveat that interactions identified by two-hybrid experiments are not always fully representative, our study indicates that MamJ possesses two distinct protein-protein interaction domains: one located in the C-terminal region and a second one in the N-terminal region. We were able to narrow down the N-terminal interaction domain to residues 23 to 81, which are located upstream of the CAR domain, while the C-terminal interaction domain encompasses residues 295 to 368. However, we assume the C-terminal interaction motif to be even smaller than 74 residues, because our complementation study suggested the Ala-rich domain (residues 293 to 334) is dispensable for MamJ function. The capacity of Ala-rich domain of binding to MamJ and MamK might arise from a few residues in its C terminus that are part of a larger interaction motif primarily spanning the sequence region between the Ala-rich and Gly-rich domains. However, both identified interaction domains show MamJ binding and thus might confer MamJ oligomerization, although the putative function of homo-oligomerization remains elusive. In addition, we found both domains interact with MamK, which would be a prerequisite for MamJ's presumed function to align magnetosomes along filamentous MamK. Although the subcellular localization of MamK was predicted to be cytoplasmic, it was recently identified as being associated with the MM by proteomic analysis of magnetosomes in Magnetospirillum magneticum (36), which might be due to tight binding to MM-associated MamJ. However, the indications presented here for a physical interaction between MamK and MamJ require additional confirmation by other techniques, such as pull-down or coimmunoprecipitation assays.
We previously observed cytoplasmic localization of MamJ-EGFP if expressed in the 40-kb deletion mutant MSR-1B, suggesting that MamK and/or other determinants encoded within the magnetosome island specifically direct the linear localization of MamJ. In contrast, intracellular localization of MamK appears to be independent from its cognate interaction partner, despite the observed interaction with MamJ. When expressed in both the ΔmamJ mutant and strain MSR-1B (data not shown), MamK-EGFP generated a linear fluorescence signal, which was consistent with the position of the magnetosome chain. An essentially identical localization of fluorescently tagged MamK was found in a ΔmamK mutant of M. magneticum and E. coli, suggesting MamK oligomerizes into a filament independently of MamJ or other MM proteins (14, 24). However, reported localization patterns of fluorescent MamK fusion proteins seem confined to the length of a magnetosome chain and do not fully extend from pole to pole, as observed, for example, for MamA-GFP or MamJ-EGFP. As this in conflict with the pole-to-pole localization of the protein filaments observed by cryo-ET, it will require further clarification (13, 15, 26).
In conclusion, our study provides further evidence that chain assembly in MTB is structurally and mechanistically complex and under subtle genetic control. The next questions to be solved are the process of magnetosome attachment to the MamK filaments and the mechanism driving the dynamic localization of magnetosomes, and it further remains to be shown whether components in addition to MamK and MamJ are involved in magnetosome chain formation.
Supplementary Material
Acknowledgments
We are grateful to Barry L. Wanner and Kirill A. Datsenko (Purdue University, West Lafayette, IN) for generously providing strain BW29427. We thank Emanuel Katzmann for help with two-hybrid studies. We are grateful to Wolfgang Heyser and Anke Toltz (University of Bremen, Germany) for support and access to the electron microscope. The continued support of Friedrich Widdel (Department of Microbiology, MPI Bremen) is greatly acknowledged.
This work was supported by the BMBF BioFuture program and the Max Planck Society.
Footnotes
Published ahead of print on 29 June 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ball, L. J., R. Kuhne, J. Schneider-Mergener, and H. Oschkinat. 2005. Recognition of proline-rich motifs by protein-protein-interaction domains. Angew. Chem. Int. Ed. 44:2852-2869. [DOI] [PubMed] [Google Scholar]
- 2.Blakemore, R. P. 1975. Magnetotactic bacteria. Science 190:377-379. [DOI] [PubMed] [Google Scholar]
- 3.Braun, V. 1995. Energy-coupled transport and signal-transduction through the Gram-negative outer-membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 4.Dove, S. L., J. K. Joung, and A. Hochschild. 1997. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 386:627-630. [DOI] [PubMed] [Google Scholar]
- 5.Flies, C. B., J. Peplies, and D. Schüler. 2005. Combined approach for characterization of uncultivated magnetotactic bacteria from various aquatic environments. Appl. Environ. Microbiol. 71:2723-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grünberg, K., E.-C. Müller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schüler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heyen, U., and D. Schüler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536-544. [DOI] [PubMed] [Google Scholar]
- 9.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 10.Karlin, S., V. Brendel, and P. Bucher. 1992. Significant similarity and dissimilarity in homologous proteins. Mol. Biol. Evol. 9:152-167. [DOI] [PubMed] [Google Scholar]
- 11.Kirschvink, J. L. 1982. Paleomagnetic evidence for fossil biogenic magnetite in western Crete. Earth Planet. Sci. Lett. 59:388-392. [Google Scholar]
- 12.Kobayashi, A., J. L. Kirschvink, C. Z. Nash, R. E. Kopp, D. A. Sauer, L. E. Bertani, W. F. Voorhout, and T. Taguchi. 2006. Experimental observation of magnetosome chain collapse in magnetotactic bacteria: sedimentological, paleomagnetic, and evolutionary implications. Earth Planet. Sci. Lett. 245:538-550. [Google Scholar]
- 13.Komeili, A. 2006. Cell biology of magnetosome formation, p. 163-174. In D. Schüler (ed.), Magnetoreception and magnetosomes in bacteria. Springer, Heidelberg, Germany.
- 14.Komeili, A., Z. Li, D. Newman, and G. Jensen. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242-245. [DOI] [PubMed] [Google Scholar]
- 15.Komeili, A., H. Vali, T. J. Beveridge, and D. Newman. 2004. Magnetosome vesicles are present prior to magnetite formation and MamA is required for their activation. Proc. Natl. Acad. Sci. USA 101:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladant, D., and G. Karimova. 2000. Genetic systems for analyzing protein-protein interactions in bacteria. Res. Microbiol. 151:711-720. [DOI] [PubMed] [Google Scholar]
- 17.Larsen, R. A., D. Foster-Hartnett, M. A. McIntosh, and K. Postle. 1997. Regions of Escherichia coli TonB and FepA proteins essential for in vivo physical interactions. J. Bacteriol. 179:3213-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen, R. A., G. E. Wood, and K. Postle. 1993. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol. Microbiol. 10:943-953. [DOI] [PubMed] [Google Scholar]
- 19.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 20.O'Dushlaine, C. T., E. J. Edwards, S. D. Park, and D. C. Shields. 2005. Tandem repeat copy-number variation in protein-coding regions of human genes. Genome Biol. 6:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philipse, A., and D. Maas. 2002. Magnetic colloids from magnetotactic bacteria: chain formation and colloidal stability. Langmuir 18:9977-9984. [Google Scholar]
- 22.Phizicky, E. M., and S. Fields. 1995. Protein-protein interactions: methods for detection and analysis. Microbiol. Rev. 59:94-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 24.Pradel, N., C. Santini, A. Bernadac, Y. Fukumori, and L. Wu. 2006. Biogenesis of actin-like bacterial cytoskeletal filaments destined for positioning prokaryotic magnetic organelles. Proc. Natl. Acad. Sci. USA 103:17485-17489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Scheffel, A., M. Gruska, D. Faivre, A. Linaroudis, P. L. Graumann, J. M. Plitzko, and D. Schüler. 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110-115. [DOI] [PubMed] [Google Scholar]
- 27.Scheffel, A., and D. Schüler. 2006. Magnetosomes in magnetotactic bacteria, p. 167-191. In J. M. Shively (ed.), Complex intracellular structures in prokaryotes, vol. 2. Springer, Berlin, Germany. [Google Scholar]
- 28.Schübbe, S., M. Kube, A. Scheffel, C. Wawer, U. Heyen, A. Meyerdierks, M. H. Madkour, F. Mayer, R. Reinhardt, and D. Schüler. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schübbe, S., C. Würdemann, J. Peplies, U. Heyen, C. Wawer, F. O. Glöckner, and D. Schüler. 2006. Transcriptional organization and regulation of magnetosome operons in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 72:5757-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schüler, D. 2006. Magnetoreception and magnetosomes in bacteria. Springer, Heidelberg, Germany.
- 31.Schüler, D. 2004. Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense. Arch. Microbiol. 181:1-7. [DOI] [PubMed] [Google Scholar]
- 32.Schüler, D., S. Spring, and D. A. Bazylinski. 1999. Improved technique for the isolation of magnetotactic spirilla from a freshwater sediment and their phylogenetic characterization. Syst. Appl. Microbiol. 22:466-471. [DOI] [PubMed] [Google Scholar]
- 33.Schüler, D., R. Uhl, and E. Baeuerlein. 1995. A simple light scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol. Lett. 132:139-145. [Google Scholar]
- 34.Schultheiss, D., R. Handrick, D. Jendrossek, M. Hanzlik, and D. Schüler. 2005. The presumptive magnetosome protein Mms16 is a poly(3-hydroxybutyrate) granule-bound protein (phasin) in Magnetospirillum gryphiswaldense. J. Bacteriol. 187:2416-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultheiss, D., and D. Schüler. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch. Microbiol. 179:89-94. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka, M., Y. Okamura, A. Arakaki, T. Tanaka, H. Takeyama, and T. Matsunaga. 2006. Origin of magnetosome membrane: proteomic analysis of magnetosome membrane and comparison with cytoplasmic membrane. Proteomics 6:5234-5247. [DOI] [PubMed] [Google Scholar]
- 37.Ullrich, S., M. Kube, S. Schübbe, R. Reinhardt, and D. Schüler. 2005. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J. Bacteriol. 187:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verstrepen, K. J., A. Jansen, F. Lewitter, and G. R. Fink. 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37:986-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williamson, M. P. 1994. The structure and function of proline-rich regions in proteins. Biochem. J. 297:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.