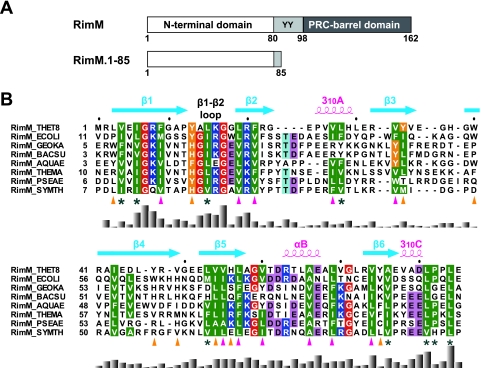
FIG. 1.
(A) Predicted domain structure of the bacterial RimM proteins (top) and the RimM fragment used for our structural and biochemical studies (bottom). Residue numbers are for the RimM protein from T. thermophilus. The two conserved tyrosine residues are indicated by the one-letter amino acid code. (B) Amino acid sequence alignment of residues 1 to 85 in T. thermophilus RimM from different bacterial species. The positions of secondary structure elements, as observed in RimM.1-85 from T. thermophilus, are shown above the sequence. The side chains of the conserved hydrophobic residues involved in the core and on the side surfaces of the β-barrel are marked below with magenta or orange triangles, respectively. Conserved but solvent-exposed hydrophobic residues are marked with asterisks. Conserved glycine, hydrophobic, and aromatic residues are shown in red, green, or orange, respectively. Positively and negatively charged conserved residues are highlighted in blue or pink, respectively. The multiple sequence alignment was performed using ClustalX (7) and was adjusted manually to align the structurally significant residues. The histogram below the sequence indicates degrees of similarity. Species abbreviations: THET8, Thermus thermophilus HB8; SYMTH, Symbiobacterium thermophilum; AQUAE, Aquifex aeolicus; THEMA, Thermotoga maritima; GEOKA, Geobacillus kaustophilus; BACSU, Bacillus subtilis; ECOLI, Escherichia coli; PSEAE, Pseudomonas aeruginosa.