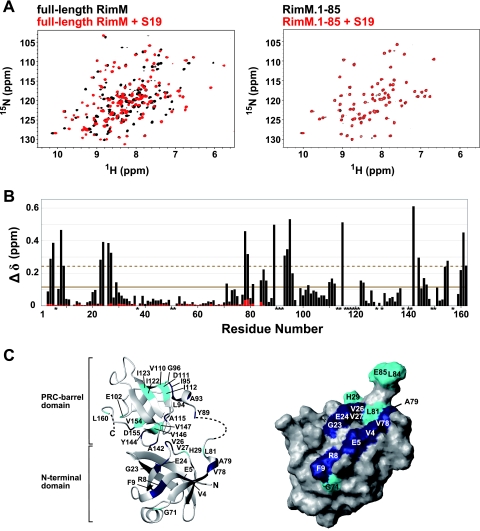
FIG. 8.
NMR studies of full-length RimM and RimM.1-85 with S19. (A) 1H-15N HSQC spectra of free full-length RimM (black) and the complex with S19 (red) obtained at 45°C (left) and spectra of free RimM.1-85 (black) and its 1:1 molar mixture with S19 (red) monitored at 45°C (right). (B) Chemical shift changes of full-length RimM (black bars) and RimM.1-85 (red bars) observed by the addition of S19. The chemical shift change, Δδ, was determined as follows: Δδ = [(ΔδHN)2 + (ΔδN/6.5)2]1/2 (29), where ΔδHN and ΔδN are the chemical shift differences for HN and 15N, respectively. The mean value is shown by a continuous line; the mean value plus 1 standard deviation is shown by a dashed line. Asterisks indicate residues with 1H-15N resonances that were not assigned in the S19-free or S19-bound form of full-length RimM. (C) Mapping of perturbed residues on the homology-modeled structure of full-length RimM (left) and the surface of RimM.1-85, which is presented in the same orientation as in Fig. 3B (right). The 3D model of full-length RimM from T. thermophilus was obtained using the homology modeling approach with the SWISS-MODEL Protein Modeling Server (http://swissmodel.expasy.org/) (36) on the basis of the crystal structure of P. aeruginosa RimM. Disordered regions in the crystal structure are depicted by a dashed line. Residues with chemical shift changes above average are cyan, and residues with chemical shift changes above the mean value plus 1 standard deviation are navy.