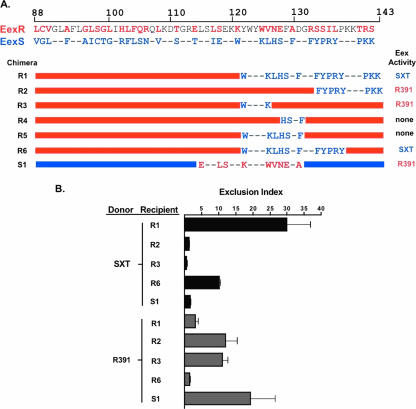
FIG. 1.
The amino acid residues that determine EexS and EexR exclusion specificities are located in distinct regions of the proteins. (A) Schematic depiction of the Eex chimeric constructs and their exclusion activities. The top two lines show the last 56 aa of EexR and EexS. Blue and red letters represent residues specific to EexS or EexR, respectively. Dashes represent conserved residues. In the maps of the chimeras, red and blue bars represent EexR and EexS sequences, respectively. (B) Exclusion activities of recipients expressing the indicated chimeric Eex proteins are expressed as exclusion index scores as previously described (17). Bacterial conjugation assays were performed as previously described (17). All recipients were E. coli CAG18439 derivatives expressing the indicated chimeric exclusion protein. The E. coli donors were MG1655 SXT ΔfloR::kan and MG1655 R391. Each bar represents the mean from three experiments. Error bars, standard deviations.