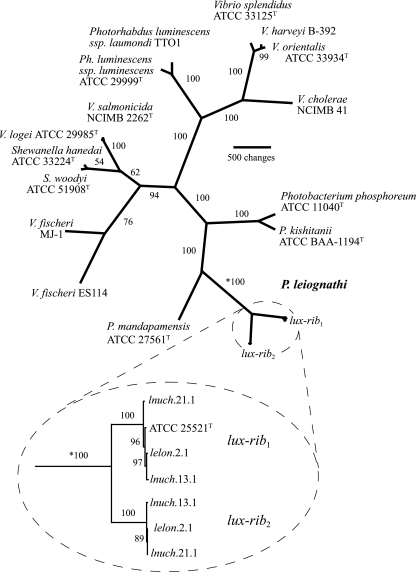
FIG. 3.
Phylogenetic hypothesis of relationships among luminous bacteria based on lux and rib gene sequences. The tree is unrooted because bacterial lux genes are present only in members of the Vibrionaceae, Shewanellaceae, and Enterobacteriaceae; no outgroup bearing the lux genes is known. The total number of aligned nucleotide positions in the data set is 11,021; exclusion of noncoding spacer regions and parsimony-uninformative characters resulted in 4,745 nucleotides for analysis. The single most parsimonious hypothesis is shown (length, 13,739; consistency index, 0.615; retention index, 0.712). Numbers at nodes are jackknife resampling values. The two lux-rib operons from P. leiognathi strains lnuch.13.1, lelon.2.1, and lnuch.21.1 (circled with dashed line) have distinct sequences but are each other's closest relatives (circled inset below the main figure; the asterisk indicates that the jackknife value is for the same branch on each part of the figure). The primary lux-rib operon (lux-rib1) is proximal to putA in all strains, whereas the secondary lux-rib operon (lux-rib2) is flanked by putative bacterial transposases in strain lnuch.13.1. Phylogenetic analysis based on housekeeping genes (i.e., gapA, gyrB, recA, rpoA, and rpoD) of Photobacterium species yielded trees consistent with the lux-rib hypothesis shown here.