Abstract
The sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough possesses four periplasmic hydrogenases to facilitate the oxidation of molecular hydrogen. These include an [Fe] hydrogenase, an [NiFeSe] hydrogenase, and two [NiFe] hydrogenases encoded by the hyd, hys, hyn1, and hyn2 genes, respectively. In order to understand their cellular functions, we have compared the growth rates of existing (hyd and hyn1) and newly constructed (hys and hyn-1 hyd) mutants to those of the wild type in defined media in which lactate or hydrogen at either 5 or 50% (vol/vol) was used as the sole electron donor for sulfate reduction. Only strains missing the [Fe] hydrogenase were significantly affected during growth with lactate or with 50% (vol/vol) hydrogen as the sole electron donor. When the cells were grown at low (5% [vol/vol]) hydrogen concentrations, those missing the [NiFeSe] hydrogenase suffered the greatest impairment. The growth rate data correlated strongly with gene expression results obtained from microarray hybridizations and real-time PCR using mRNA extracted from cells grown under the three conditions. Expression of the hys genes followed the order 5% hydrogen > 50% hydrogen > lactate, whereas expression of the hyd genes followed the reverse order. These results suggest that growth with lactate and 50% hydrogen is associated with high intracellular hydrogen concentrations, which are best captured by the higher activity, lower affinity [Fe] hydrogenase. In contrast, growth with 5% hydrogen is associated with a low intracellular hydrogen concentration, requiring the lower activity, higher affinity [NiFeSe] hydrogenase.
Hydrogen is intimately involved in the metabolism of sulfate-reducing bacteria (SRB) of the genus Desulfovibrio (17). In addition to utilizing molecular hydrogen directly as an electron donor for sulfate reduction, hydrogen may play a central role as an intermediate in the generation of a chemiosmotic gradient from the oxidation of organic molecules. This process has become known as hydrogen cycling (18). The current emphasis on developing more efficient energy production strategies has made understanding SRB metabolism a critical endeavor, especially with respect to the SRB connection with hydrogen and the impact of SRB on the petroleum industry through oil reservoir souring and pipeline corrosion.
Hydrogenases catalyze the heterolytic cleavage of molecular hydrogen into protons and electrons (H2 ↔ 2H+ + 2e) (9). These enzymes are present ubiquitously in both Archaea and Bacteria, including many SRB. The fully sequenced Desulfovibrio vulgaris Hildenborough has a total of six hydrogenases (14). Four of them are periplasmic and therefore presumably are involved in hydrogen oxidation, including a soluble iron-only [Fe] hydrogenase (Hyd) (16), two membrane-associated nickel-iron [NiFe] hydrogenase isozymes (Hyn1 and Hyn2) (14, 23), and a membrane-associated nickel-iron-selenium [NiFeSe] hydrogenase (Hys) (31). The [NiFe] hydrogenases are widely distributed in SRB (35), but many Desulfovibrio spp. are exceptional in also possessing a soluble [Fe] hydrogenase. Despite the importance of these enzymes, it remains unclear why D. vulgaris possesses four periplasmic hydrogenases when a single enzyme could be sufficient.
Several hypotheses have been proposed to explain the D. vulgaris hydrogenase redundancy. Possessing multiple hydrogenases may allow D. vulgaris to cope with (i) changes in metal availability (6, 30), (ii) exposure to hydrogenase inhibitors, or (iii) varying environmental concentrations of molecular hydrogen (35). Alternatively, specific hydrogenases may function in separate metabolic pathways (13, 34). Since three of the hydrogenases have dissimilar metal compositions in their active sites and consequently possess unique kinetic properties, the effect of altering hydrogen concentration on in vivo hydrogenase utilization needs to be considered. It has been demonstrated that the [Fe] hydrogenase has the highest specific activity (50,000 μmol min−1 mg protein−1) and the lowest binding affinity (Km = 100 μM) for hydrogen uptake (7), whereas the [NiFe] and [NiFeSe] hydrogenases have lower specific activities (100 to 1,000 μmol min−1 mg protein−1) and higher binding affinities (Km = 1 μM) for hydrogen uptake (23, 31, 32). Consequently, it would be advantageous for D. vulgaris to utilize [Fe] hydrogenase preferentially at high hydrogen partial pressures and the nickel-containing hydrogenases at low hydrogen partial pressures.
To test this hypothesis, cultures of the wild-type strain and mutant strains with deletions of either the hyd, hyn1, hys, or both the hyn1 and hyd genes were grown with high and low concentrations of hydrogen or lactate as the electron donor for sulfate reduction. In addition to growth rate determination, RNA was extracted from the wild-type cells to assess the gene expression patterns under these conditions. Both the growth and expression data suggest that the [NiFeSe] hydrogenase is important for growth at lower concentrations of hydrogen, while the [Fe] hydrogenase facilitates rapid growth at higher hydrogen concentrations and when lactate is used as the electron donor.
MATERIALS AND METHODS
Materials.
Gasses, including H2, CO2, and N2, were obtained from Praxair Products, Inc., Edmonton, Alberta, Canada. Trizol, Bacterial Max reagent, the SuperScript indirect cDNA labeling system, the BioPrime Plus array CGH genomic labeling system, Alexa Fluor 555 and 647, sodium dodecyl sulfate, standard saline citrate (SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]), herring sperm DNA, and SYBR GreenER two-step qRT-PCR kits were obtained from Invitrogen, Burlington, Ontario, Canada. RNeasy kits and RNase-free DNase were from QIAGEN, Mississauga, Ontario, Canada. G418 sulfate was purchased from Calbiochem. Deoxyoligonucleotide primers were synthesized by University Core DNA Services, University of Calgary, Calgary, Alberta, Canada.
Media and culture conditions.
D. vulgaris was cultured at 30°C in 1-liter Erlenmeyer flasks containing 500 ml of defined Widdel-Pfenning (WP) medium containing 3 mM acetate and 28 mM sulfate (34, 36). This medium contains added nickel and selenite, which are required for expression of [NiFe] and [NiFeSe] hydrogenases (30). The medium was bubbled with oxygen-free mixed gas consisting of either 0.5, 5, or 50% (vol/vol) hydrogen and 10% CO2, and the balance made up by N2. Alternatively, 65 mM lactate was the sole electron donor in cultures gassed with 10% CO2 and 90% N2. Gas was delivered at 100 ml/min after being passed through a heated (325°C) copper reduction tube (Thermolyne 21100 tube furnace; VWR International, Mississauga, Ontario, Canada) for oxygen removal and through a water bottle for humidification. Gas was mixed from pure H2, CO2, and N2 with Sierra model C100L gas mixers from GS HighTech Controls Inc., Calgary, Alberta, Canada.
Construction of mutant strains.
The 500-bp regions located upstream and downstream of hysBA (DVU1917 and DVU1918) were PCR amplified using primer pairs p300f/p302r and p354f/p355r, respectively (Table 1). The amplified fragments were cloned sequentially into pNOT19 by digestion with SacI and BamHI as well as HindIII and BamHI and ligation to obtain pNOTΔhys. The latter was cleaved with BamHI and ligated to the cat gene-containing 1.4-kb BamHI fragment from pUC19Cm to obtain pNOTΔhysCm. NotI digestion of the latter and insertion of the 4.2-kb NotI fragment from pMOB2 gave pNOTΔhysCmMob, which was transformed into Escherichia coli S17-1. Following conjugation of E. coli S17-1(pNOTΔhysCmMob) and D. vulgaris, single-crossover integrants were obtained on medium E plates containing kanamycin (Km) and chloramphenicol (Cm), as described elsewhere (10). Southern blot analysis with a probe, generated by PCR amplification with primers p354f and p355r as described elsewhere (10), indicated insertion downstream from hysA. Growth of the integrant in defined medium containing Cm and 5% (wt/vol) sucrose (Suc), as described by Pohorelic et al. (21), produced a double-crossover mutant, in which the hysBA coding regions were replaced with the cat gene.
TABLE 1.
Bacterial strains, plasmids, vectors and primers used
| Strain, plasmid, or primer | Genotype, comment(s), and/or source or referencea |
|---|---|
| Strain | |
| D. vulgaris subsp. vulgaris strain Hildenborough | NCIMB 8303; isolated from clay soil near Hildenborough, United Kingdom (22) |
| D. vulgaris Hyd100 | Δhyd; Sucr Cmr (21) |
| D. vulgaris NiFe100 | Δhyn1; Sucr Cmr (11) |
| D. vulgaris Hys100 | Δhys; Sucr Cmr (this study) |
| D. vulgaris HydHyn1 | Δhyd Δhyn1; Sucr Cmr Kmr (this study) |
| E. coli S17-1 | thi pro hsdR hsdM+recA RP4-2 (Tc::Mu, Km::Tn7) (26) |
| Plasmid | |
| pUC19Cm | pUC19 containing the Cm gene (10) |
| pNOT19 | Cloning vector pUC19; NdeI site replaced by a NotI site (25) |
| pMOB2 | Contains oriT of plasmid RP4 and Bacillus subtilis sacBR genes on a 4.5-kb NotI fragment; Kmr Cmr (25) |
| pBSL180 | Source of nptII gene (1) |
| pNOT180 | 1.3-kb SacI fragment containing nptII cloned in pNOT19 (this study) |
| pNOTΔNiFeCm | 11 |
| pNOTΔNiFeKm, pNOTΔNiFeKmMob | This study |
| pNOTΔhys, pNOTΔhysCm, pNOTΔhysCmMob | This study |
| Primer | |
| p225f | 5′tcgaaagcttCGGCGCGGTAACACGATT (+3259 to +3276) |
| p228r | 5′agtacggatccCATCGTGTGGTTGGCGAC (+4007 to +3991) |
| p293r | 5′TGATGTAATCGCCAAGGGCCG (+7297 to +7277) |
| p292f | 5′GGCGATACTCGCTTCGTT (+3237 to +3254) |
| p354f | 5′-tcgaggatccTGCTGCCGACGCCCATGATGT (+2593 to +2613) |
| p355r | 5′-tcgagagctcTGCCACCCAGCATCTCTGCCA (+3112 to +3092) |
| p356r | 5′-CATGCCTAGCACATGCATGGT (+3141 to +3121) |
| p300f | 5′-tcgaaagcttGGGCATGACAGATTGACCAC (−535 to −516) |
| p302r | 5′-tcgaggatccAGTGCCAGCCAATAGAGTGAA (−27 to −47) |
| p304f | 5′-GGCGTGTCCTACAACATCATC (−624 to −604) |
For the primers, the positions given are relative to the start of the hysB coding region (bp +1 to +953); hysA is located from +987 to +2519, hynB1 is located from +3576 to +4581, and hynA1 is located from +4581 to +6281. Lowercase letters are bases added to create restriction endonuclease cleavage sites.
A hyn1 hyd double mutant was constructed by introducing the hyn1 mutation into D. vulgaris Hyd100. Because this strain is already Cm resistant through replacement mutagenesis of the hyd genes, we used the nptII gene, encoding Km resistance, as a second selectable marker. Although D. vulgaris has high endogenous Km resistance, it is sensitive to G418, a related antibiotic (2). Hence, G418 was used in all selections by adding 20 μl of a filter-sterilized solution of 30 mg/ml per ml of liquid medium or by spreading 300 μl of the same onto medium E-Cm plates (∼25 ml). The nptII gene was retrieved from pBSL180 by digestion with SacI and was ligated to SacI-digested pNOT19, giving pNOT180. Plasmid pNOTΔNiFeCm was digested with BamHI and ligated to the nptII gene-containing 1.3-kb BamHI fragment from pNOT180, giving pNOTΔNiFeKm. NotI digestion of the latter and insertion of the 4.2-kb NotI fragment from pMOB2 gave pNOTΔNiFeKmMob, which was transformed into E. coli S17-1. Following conjugation of E. coli S17-1 (pNOTΔNiFeKmMob) and D. vulgaris Hyd100, single-crossover integrants were obtained on medium E plates containing Cm and G418. Southern blot analysis using a probe, obtained by PCR amplification with primers p225f and p228r and 32P-labeling, indicated that all of the integrants were insertions downstream from hynA1. Growth of one of these, D. vulgaris HH17, in defined medium containing Cm, G418, and 5% (wt/vol) Suc resulted in a double-crossover mutant, D. vulgaris Hyn1-Hyd, which had all of hynB1 and most of hynA1 replaced with the nptII gene.
Growth analysis.
Samples (1 ml) were withdrawn periodically from growing cultures through a butyl-rubber-stopper-closed sampling port to determine the optical density at 600 nm (OD600) or other metabolite concentrations. In some experiments, sulfate, acetate, and lactate concentrations were determined using high-pressure liquid chromatography, as previously described (15). Replicate growth curve data (from a minimum of four trials) from the wild type and the four mutant strains were analyzed by drawing a logarithmic line of best fit through the exponential-growth-phase data points for each trial and using the slope to calculate the specific growth rate constant. Significant differences in growth rate or maximum cell density were inferred when the P value was less than 0.05 from a one-way analysis of variance (ANOVA) with a Bonferroni's post hoc test. If the ANOVA test declared that the data for one or more of the strains being compared were significantly different, a post hoc test was conducted with the data sets for each of the strains to determine which of the strains were growing at different rates. Since this required multiple tests, the chance of a false positive increased, and a Bonferroni's adjustment was used to maintain the false-positive error rate at 0.05. Statistical analysis was conducted with GraphPad Prism (GraphPad Software Inc., San Diego, CA).
RNA and DNA extraction and labeling.
Total RNA was extracted from mid-log-phase cells using Invitrogen Trizol and Bacterial Max reagents. The RNA was purified with the QIAGEN RNA easy kit with on-column DNase digestion. RNA quality and quantity were assessed using the ratio of absorbances at 260 and 280 nm (A260/A280), denaturing agarose gel electrophoresis with ethidium bromide staining, and with a Qubit fluorometer (Invitrogen, Burlington, Ontario, Canada). Genomic DNA (gDNA) was isolated as described by Voordouw et al. (35). Amino-modified cDNA was generated from 10 μg of total RNA using the Invitrogen SuperScript indirect cDNA labeling system and random hexamers. Amino-modified gDNA was generated using 1 μg of gDNA with the Invitrogen BioPrime Plus array CGH genomic labeling system. The amino-modified cDNA was then coupled with Alexa Fluor 647, and the amino-modified gDNA was coupled with Alexa Fluor 555.
Microarray hybridization, washes, and scans.
Dried probes from the treatment condition (cDNA) and the universal reference (gDNA) were mixed and diluted to 40 μl by using a hybridization solution containing 50% (vol/vol) formamide, 5× SSC, 0.1% (wt/vol) sodium dodecyl sulfate, and 0.1 mg of herring sperm DNA/ml. This solution then was incubated at 98°C for 5 min and then kept at 60°C. Sixteen microliters of 3× SSC solution was added to the wells at the ends of the microarray slides to maintain proper chamber humidity, and then the hybridization solution was applied to a Corning UltraGAPS microarray slide, which had been prewashed according to the manufacturer's specification (Corning, NY). These microarrays contained 70-mer oligonucleotide probes for 3,368 of the 3,394 protein-coding sequences of the D. vulgaris chromosome and were constructed as previously described (3). The slides were incubated for 17 h in Corning hybridization chambers, washed according to the manufacturer's specifications, and dried in a centrifuge. The hybridized slides were scanned on a PerkinElmer Scanarray 5000 scanner. Spot and background intensities were quantified using the Spotfinder software of The Institute for Genomic Research (TIGR) (24). The gDNA channel was used as the control for the immobilized probe concentration, spot uniformity, and labeling efficiency, as described by Talaat et al. (27) and Chhabra et al. (3). Spots were flagged and rejected if they were saturated, of irregular size, missing from one channel, or had a poor signal-to-noise ratio (less than two times the median local background level). A complete microarray analysis used RNA extracted from two independent biological samples that were hybridized to a minimum of four arrays that had duplicate spots. Data below the threshold of a low-pass filter were rejected. The remaining microarray hybridization intensities were normalized using total intensity normalization and then averaged for the duplicate spots and the four arrays. Hybridization intensity ratios, Ri, were calculated from the normalized, averaged cDNA and gDNA hybridization intensities (designated <cDNAi> and <gDNAi>, respectively), as Ri = <cDNAi>/<gDNAi>.
Differentially expressed genes were discovered with significance analysis of microarrays using the Stanford tools package (29), which averaged the normalized intensities for each gene, i, and calculated Ri. An expression ratio, RC,I, for gene i for two different conditions, A and B, was calculated as RC,i = Ri,A/Ri,B. Genes were determined to be differentially expressed if they had at least a twofold-changed RC,i and a q value of less than 5%. The q value measures the percentage of false versus significant calls for the twofold expression change. Functional categories for global analysis were defined by COGs (clusters of orthologous genes) (28). The bootstrap-supported hierarchical clustering tree was created using TIGR's multiple experiment viewer from genes identified as significant by a one-way ANOVA with adjusted Bonferroni's correction at the 1% confidence level (24). The clusters were created using Euclidean distances and average linkages.
RT-PCR.
Primers were designed to generate a product smaller than 200 bp for the hydA, hysA, and 16S rRNA genes (hydA, 5′-CTGGCAGAAGTATGCCGAGA and 5′-GGACGCAACCCTTCGTACTT; hysA, 5′-GAAATCAAGGCTGGCGAAGA and 5′-ACCACGCTGTCCCATATCGT; 16S rRNA, 5′-ATGAAGGTCCTCGGATCGTA and 5′-CCTGACTTACCAAGCAGCCTAC). The product obtained with primers for 16S rRNA was used as a loading control. The cDNA template for the real-time PCR (RT-PCR) was generated using the SYBR GreenER Two-Step qRT-PCR kit from 1 μg of total RNA. The RT-PCR was conducted in an iCycler thermal cycler in accordance with the manufacturer's specifications (Bio-Rad Laboratories Ltd, Mississauga, Ontario, Canada). The measured, 16S rRNA-normalized expression level for RNA extracted from cells grown with lactate was used as a reference for the 50% hydrogen and 5% hydrogen conditions.
Accession number.
The microarray data presented in this paper have been entered into the NCBI GEO data bank (5) under accession number GSE 8137.
RESULTS
Construction of hydrogenase mutants.
Although [NiFeSe] hydrogenase recently has been shown to be the major hydrogenase of D. vulgaris in media to which selenium was added under a variety of growth conditions (30), the hysBA genes nevertheless were easily deleted. Southern blotting using a p354f- and p355r-amplified, radiolabeled probe gave hybridizing SalI fragments of 2.5 kb, of 8.3 and 1.2 kb, and of 6.5 kb for the wild type, the downstream integrant, and the replacement mutant strain, respectively (data not shown). Use of primers p304f and p356r yielded a 2.5-kb product for D. vulgaris Hys100, confirming the replacement of its hys genes by the cat gene. Construction of a hyn1 hyd double mutant that was resistant to both Cm and G418 likewise was confirmed by Southern blotting. A p225f- and p228r-amplified radiolabeled probe hybridized with fragments of 6.6 kb, of 4.1 and 5.9 kb, and of 0.8 kb for the wild type, the downstream integrant, and the replacement mutant strain, respectively. Use of primers p292f and p293r gave a 2.4-kb PCR product for D. vulgaris Hyn1-Hyd, confirming replacement of its hyn1 genes by nptII.
Physiological analysis.
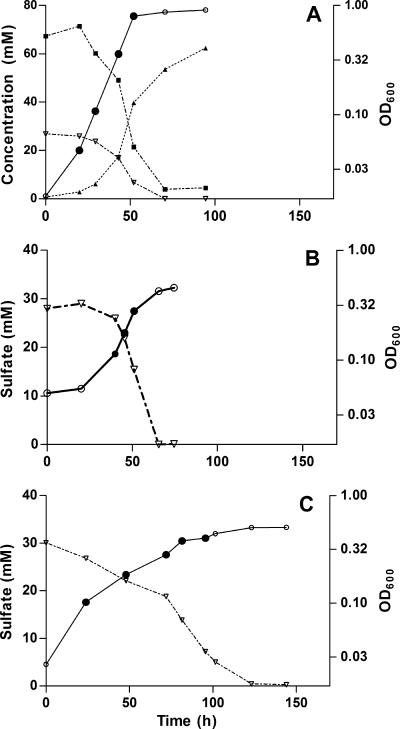
Typical growth curves for D. vulgaris grown in the culture system using WP medium with lactate, 50% (vol/vol) hydrogen, or 5% (vol/vol) hydrogen as the sole electron donor are shown in Fig. 1. The utilization of sulfate (Fig. 1), the utilization of lactate, and the production of acetate (Fig. 1A) also are shown, although these were not determined routinely. Despite the intrinsic importance of hydrogenases to D. vulgaris metabolism, strains missing the Hyd, Hyn1, or Hys hydrogenase or both the Hyn1 and Hyd hydrogenases were able to grow with either lactate or hydrogen as the sole electron donor for sulfate reduction. When the D. vulgaris strains were grown with lactate as the sole electron donor, strains lacking [Fe] hydrogenase were the most affected. The hyd and hyn1 hyd mutants grew significantly more slowly than the wild type (Fig. 2A) and reached a lower stationary-phase cell density (Fig. 2B).
FIG. 1.
Growth curves for wild-type D. vulgaris in WP medium with lactate (A), 50% (vol/vol) hydrogen (B), or 5% (vol/vol) hydrogen (C) as the sole electron donor for sulfate reduction. The linear scale on the left corresponds to the following, each as a function of time: ▿, sulfate concentration (A to C); ▪ concentration of lactate (A); and ▴, concentration of acetate (A). The logarithmic scale on the right indicates the cell density at OD600 (○ and •). The filled circles represent data used for calculation of the specific growth rate constant, k.
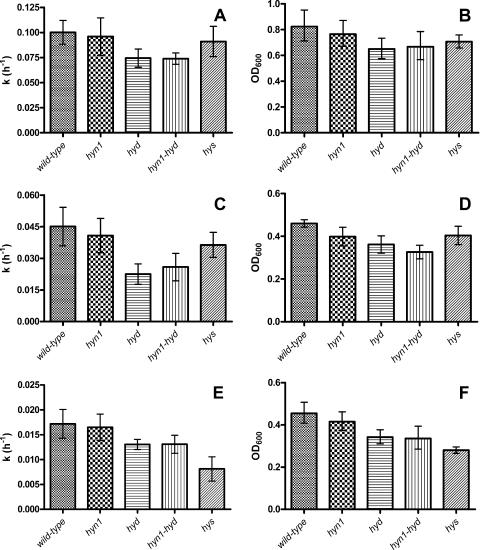
FIG. 2.
Specific growth rate constants (k) and stationary-phase cell densities (OD600) of wild-type and mutant strains grown with lactate (A and B), 50% (vol/vol) hydrogen (C and D), or 5% (vol/vol) hydrogen (E and F) as the sole electron donor for the reduction of sulfate. The data are averages from at least four independent experiments. Error bars represent 95% confidence intervals.
When the strains were grown with 50% (vol/vol) hydrogen as the sole electron donor, the specific growth rate constant (k) of the wild-type strain decreased from k = (0.100 ± 0.011) h−1 for growth with lactate to k = (0.045 ± 0.010) h−1 for growth with 50% hydrogen, with 0.011 and 0.010 h−1 being the standard deviations from the means, respectively. Strains lacking the hyd genes again experienced the most impaired growth characteristics (Fig. 2C and D).
Lowering the hydrogen concentration to 5% (vol/vol) gave a further reduction in the specific growth rate constant of the wild type, to k = (0.017 ± 0.004) h−1, whereas that of the hys strain was lowered even more, to k = (0.008 ± 0.002) h−1 (Fig. 2E). The hys strain also had the lowest stationary-phase cell density, as shown in Fig. 2F. Note that although the wild-type and mutant cells grew significantly more slowly at 5% than at 50% hydrogen, with the exception of the hys strain they reached the same stationary-phase cell density.
The wild-type, hyn1, and hyd strains also were grown with 0.5% (vol/vol) hydrogen gas (data not shown). Under these conditions, growth was extremely slow and difficult to maintain, and it produced insufficient RNA for microarray analysis. The specific growth rate constant for the wild type was estimated at k = (0.0049 ± 0.0004) h−1.
Analysis of global gene expression.
RNA extracted from mid-log-phase cultures of wild-type D. vulgaris, grown with either 65 mM lactate, 50% hydrogen, or 5% hydrogen as the sole electron donor, was analyzed using full-genome microarrays (3). For the global analysis, genes were classified as differentially expressed if they exhibited an expression ratio change of greater than twofold and possessed a q value of zero (0% chance of false discoveries). The greatest change in gene expression was between the cells grown with 5% hydrogen and the cells grown with lactate (604 genes; 343 up and 261 down) and between cells grown with 50% hydrogen and cells grown with lactate (637 genes; 427 up and 210 down). Comparing cells grown with both hydrogen concentrations to those grown with lactate, only 266 genes (158 up, 108 down) had a twofold expression change. Likewise, for cells grown with 5% and with 50% hydrogen, fewer genes with changed expression were observed than were observed in a comparison with growth with lactate (263 genes; 77 up and 186 down).
As defined by COG categories, D. vulgaris cells utilizing hydrogen as the sole electron donor exhibited increased gene expression relative to cells utilizing lactate in the categories vii (posttranslational modification, protein turnover, and chaperones), xv (amino acid transport and metabolism), and xxii (function unknown), in which the largest number of genes was upregulated (Table 2). Genes in category xiii (energy production and conversion) exhibited a similar number of up- and downregulated genes (Table 2), whereas genes in category i (translation, ribosomal structure, and biogenesis) were downregulated, reflecting the lower growth rates with hydrogen (Fig. 2C and E) than with lactate (Fig. 2A). Relative to cells grown with 50% hydrogen, those grown with 5% hydrogen showed downregulation of genes in COG categories i (translation, ribosomal structure, and biogenesis), xv (amino acid transport and metabolism), xix (inorganic ion transport and metabolism), and xxii (function unknown), which contained the greatest number of downregulated genes. Genes in category xiii (energy production and conversion) again appeared subject to compensatory changes (Table 2).
TABLE 2.
Numbers of genes up- and downregulated in each of 22 COG categories in D. vulgaris wild-type cells grown in two different conditions
| COG functional category | No. of genes upregulated (Up) or downregulated (Down) under the following condition:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 5% H2 vs lactate (Up) | 5% H2 vs lactate (Down) | 50% H2 vs lactate (Up) | 50% H2 vs lactate (Down) | H2 vs lactate (Up) | H2 vs lactate (Down) | 5% H2 vs 50% H2 (Up) | 5% H2 vs 50% H2 (Down) | |
| i. Translation, ribosomal structure, and biogenesis | 4 | 45 | 4 | 25 | 2 | 25 | 1 | 8 |
| ii. RNA processing and modification | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| iii. Transcription | 3 | 3 | 6 | 4 | 3 | 2 | 0 | 1 |
| iv. DNA replication, recombination, and repair | 6 | 2 | 4 | 4 | 1 | 1 | 2 | 3 |
| v. Cell division and chromosome partitioning | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| vi. Defense mechanisms | 1 | 2 | 2 | 2 | 1 | 0 | 0 | 1 |
| vii. Posttranslational modification, protein turnover, and chaperones | 12 | 10 | 19 | 4 | 11 | 3 | 2 | 4 |
| viii. Cell envelope biogenesis and outer membrane | 11 | 5 | 15 | 6 | 3 | 2 | 4 | 3 |
| ix. Cell motility and secretion | 9 | 1 | 10 | 1 | 5 | 1 | 1 | 3 |
| x. Signal transduction mechanisms | 14 | 7 | 18 | 4 | 6 | 2 | 3 | 3 |
| xi. Extracellular structures | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| xii. Intracellular trafficking, secretion, and vesicular transport | 2 | 5 | 4 | 2 | 0 | 2 | 0 | 3 |
| xiii. Energy production and conversion | 32 | 28 | 28 | 25 | 13 | 14 | 14 | 16 |
| xiv. Carbohydrate transport and metabolism | 0 | 4 | 10 | 3 | 0 | 0 | 0 | 2 |
| xv. Amino acid transport and metabolism | 21 | 10 | 31 | 9 | 8 | 3 | 7 | 11 |
| xvi. Nucleotide transport and metabolism | 2 | 3 | 3 | 1 | 0 | 1 | 0 | 3 |
| xvii. Coenzyme metabolism | 7 | 6 | 10 | 4 | 6 | 2 | 1 | 3 |
| xviii. Lipid metabolism | 4 | 3 | 2 | 4 | 1 | 1 | 1 | 2 |
| xix. Inorganic ion transport and metabolism | 5 | 4 | 15 | 3 | 1 | 0 | 0 | 13 |
| xx. Secondary metabolites biosynthesis, transport, and catabolism | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| xxi. General function prediction only | 24 | 13 | 31 | 8 | 8 | 3 | 6 | 10 |
| xxii. Function unknown | 148 | 89 | 170 | 84 | 75 | 38 | 31 | 73 |
| Totala | 343 | 261 | 427 | 210 | 158 | 108 | 77 | 186 |
The overall total does not count genes belonging to multiple categories more than once.
The premise that gene expression patterns during growth on 5% and 50% hydrogen are more closely related to each other than to that during growth with lactate, as suggested by the total number of genes changed (Table 2), also is supported by the TIGR multiple-experiment-viewer-generated bootstrapped tree, in which the expression patterns of cells grown with 5 and 50% hydrogen cluster together, separately from those grown with lactate (results not shown).
Analysis of expression of specific genes.
The expression ratios of several genes of interest, including those involved in hydrogen and lactate metabolism, are displayed in Table 3. Major changes in hydrogenase gene expression were observed between cells grown in medium in which lactate or hydrogen (50% or 5%, vol/vol) is the electron donor. A shift from lactate to hydrogen causes upregulation of the hys genes and downregulation of the hyd genes. These changes are among the highest levels of all of gene expression changes observed for the entire genome. In comparing cells grown with 5% hydrogen to those grown with lactate, the hysB gene is the second most upregulated gene, whereas the hydA gene is the second most downregulated gene (Table 3). When the normalized, averaged expression ratio, Ri, is used to rank the genes in order of decreasing expression level, hysB and hysA rank 7th and 13th (hence, they are highly expressed), whereas hydB and hydA rank 1,610th and 2,469th, respectively. In contrast, during growth with lactate, hysB and hysA rank 546th and 633rd, whereas hydB and hydA rank 153rd and 111th, respectively (see Table S1B in the supplemental material). Other significant changes between growth on 5% and/or 50% hydrogen and growth on lactate include upregulation of the Ech hydrogenase operon DVU0429 to DVU0434, heterodisulfide reductase genes DVU2402 to DVU2404, nickel transport genes DVU1057 and DVU1058, iron transport genes DVU0648 and DVU0649, formate dehydrogenase genes DVU0587 and DVU0588, the gene for cytochrome c553 (DVU2797), and adjacent genes that form the operon for the Rnf electron transport complex, DVU2791 to DVU2796 (Table 3). Many of these genes were more upregulated in 5% than in 50% hydrogen relative to expression in lactate. Genes for the Qmo transmembrane complex (DVU848 to DVU850), for the Dsr transmembrane complex (DVU1286 to DVU1290), for adenylylsulfate reductase (DVU0846 and DVU0847), and for alcohol dehydrogenase (DVU2201) were downregulated relative to growth on lactate, as were many ribosomal genes (not listed in Table 3) and ATP synthase genes (DVU0774 to DVU0780). This may, in part, reflect the lower growth rates observed for 5% hydrogen compared to those for 50% hydrogen and for lactate (Fig. 2).
TABLE 3.
Selected genes that are up- or downregulated when expression levels of genes of wild-type D. vulgaris cells grown under two different conditions are compared
| Gene designation | Annotation | Gene expression ratio for cells grown under the following conditions:
|
|||||
|---|---|---|---|---|---|---|---|
| 5% H2 vs lactate
|
50% H2 vs lactate
|
5% H2 vs 50% H2
|
|||||
| Log2RC | q value (%) | Log2RC | q value (%) | Log2RC | q value (%) | ||
| DVU1917 | [NiFeSe] hydrogenase, small subunit (hysB) | 4.62 | 0.00 | 0.95 | 0.00 | 3.67 | 0.00 |
| DVU1918 | [NiFeSe] hydrogenase, large subunit (hysA) | 4.5 | 0.00 | 1.10 | 0.00 | 3.31 | 0.00 |
| DVU1769 | Periplasmic [Fe] hydrogenase, large subunit (hydA) | −4.35 | 0.00 | −2.98 | 0.00 | −1.68 | 0.00 |
| DVU1770 | Periplasmic [Fe] hydrogenase, small subunit (hydB) | −2.95 | 0.00 | −2.52 | 0.00 | −1.00 | 0.11 |
| DVU2286 | Coo hydrogenase CooM | 0.40 | 22.8 | −0.62 | 0.19 | 1.24 | 0.93 |
| DVU2287 | Coo hydrogenase CooK | −0.21 | 21.88 | −0.44 | 0.19 | 0.336 | 3.03 |
| DVU2288 | Coo hydrogenase CooL | −0.96 | 0.17 | −1.55 | 0.00 | 0.75 | 0.93 |
| DVU2289 | Coo hydrogenase CooX | 0.83 | 0.50 | −0.81 | 0.00 | 1.86 | 0.00 |
| DVU2290 | Coo hydrogenase CooU | −1.38 | 0.00 | −1.65 | 0.00 | 0.25 | 3.03 |
| DVU2291 | Coo hydrogenase CooH | 0.10 | 22.80 | −0.70 | 0.04 | 0.54 | 1.80 |
| DVU0429 | Ech hydrogenase EchF | 0.21 | 21.88 | 0.13 | 0.14 | 0.08 | 5.19 |
| DVU0430 | Ech hydrogenase EchE | 3.28 | 0.00 | 0.18 | 0.14 | 3.16 | 0.15 |
| DVU0431 | Ech hydrogenase EchD | 0.77 | 0.50 | 0.90 | 0.00 | −0.14 | 4.88 |
| DVU0432 | Ech hydrogenase EchC | 1.35 | 0.00 | 1.24 | 0.00 | 0.14 | 5.19 |
| DVU0433 | Ech hydrogenase EchB | 0.85 | 0.50 | 1.07 | 0.00 | −0.21 | 4.88 |
| DVU0434 | Ech hydrogenase EchA | 1.44 | 0.00 | 1.64 | 0.00 | −0.10 | 4.88 |
| DVU0531 | HmcF | −0.12 | 23.54 | −1.04 | 0.00 | 0.81 | 0.67 |
| DVU0532 | HmcE | −0.58 | 2.07 | −0.85 | 0.04 | 0.94 | 1.28 |
| DVU0533 | HmcD | −0.46 | 4.51 | −0.99 | 0.04 | 0.72 | 1.28 |
| DVU0534 | HmcC | −0.58 | 0.99 | −1.36 | 0.00 | 0.82 | 2.74 |
| DVU0535 | HmcB | 0.66 | 3.59 | −0.18 | 0.19 | 1.22 | 0.21 |
| DVU0536 | HmcA | 0.09 | 29.47 | −0.52 | 0.04 | 0.97 | 0.47 |
| DVU2791 | Cytochrome c family protein | 1.13 | 0.06 | −0.37 | 0.19 | 1.50 | 0.00 |
| DVU2792 | Electron transport complex protein RnfC | 0.80 | 0.33 | −0.25 | 0.19 | 1.00 | 0.21 |
| DVU2793 | Electron transport complex protein RnfD | 1.63 | 0.00 | 0.93 | 0.00 | 0.71 | 1.28 |
| DVU2794 | Electron transport complex protein RnfG | 1.05 | 0.06 | 0.17 | 0.14 | 0.83 | 0.47 |
| DVU2795 | Electron transport complex protein RnfE | 1.94 | 0.00 | 1.38 | 0.00 | 0.55 | 2.40 |
| DVU2796 | Electron transport complex protein RnfA | 1.77 | 0.00 | 0.93 | 0.00 | 0.82 | 0.93 |
| DVU0846 | Adenylyl-sulfate reductase | −1.90 | 0.00 | −1.10 | 0.00 | −0.75 | 1.28 |
| DVU0847 | Adenylyl-sulfate reductase | −1.97 | 0.00 | −0.88 | 0.04 | −1.03 | 0.11 |
| DVU0848 | QmoA | −2.16 | 0.00 | −1.43 | 0.00 | −0.71 | 1.66 |
| DVU0849 | QmoB | −2.50 | 0.00 | −1.62 | 0.00 | −0.86 | 0.67 |
| DVU0850 | QmoC | −2.90 | 0.00 | −1.73 | 0.00 | −1.26 | 0.00 |
| DVU1286 | DsrP | −1.58 | 0.00 | −1.89 | 0.00 | 0.35 | 2.74 |
| DVU1287 | DsrO | −1.53 | 0.00 | −1.60 | 0.00 | 0.10 | 5.19 |
| DVU1288 | DsrJ | −1.87 | 0.00 | −2.25 | 0.00 | 0.45 | 2.56 |
| DVU1289 | DsrK | −1.80 | 0.00 | −1.83 | 0.00 | 0.06 | 5.19 |
| DVU1290 | DsrM | −0.80 | 3.59 | −1.01 | 0.04 | 0.27 | 5.19 |
| DVU0774 | ATP synthase (atpC) | −2.05 | 0.00 | −0.18 | 0.19 | −1.86 | 0.00 |
| DVU0775 | ATP synthase (atpD) | −1.81 | 0.00 | 0.11 | 0.14 | −1.90 | 0.00 |
| DVU0776 | ATP synthase (atpG) | −1.98 | 0.00 | −0.20 | 0.19 | −1.75 | 0.00 |
| DVU0777 | ATP synthase (atpA) | −1.93 | 0.00 | −0.14 | 0.19 | −1.68 | 0.00 |
| DVU0778 | ATP synthase (atpH) | −1.56 | 0.00 | 0.49 | 0.04 | −1.97 | 0.00 |
| DVU0779 | ATP synthase F0, B | −1.14 | 0.00 | 0.96 | 0.00 | −2.08 | 0.00 |
| DVU0780 | ATP synthase F0, B′ | −1.00 | 0.10 | 0.85 | 0.00 | −1.79 | 0.00 |
| DVU1056 | NikO | −0.21 | 24.17 | 0.53 | 0.14 | −0.54 | 4.88 |
| DVU1057 | NikQ | 1.13 | 0.17 | 1.24 | 0.00 | 0.07 | 5.19 |
| DVU1058 | NikM | 0.80 | 0.33 | 1.09 | 0.00 | −0.27 | 4.88 |
| DVU2769 | NikK | −0.02 | 28.70 | 0.69 | 0.04 | −0.68 | 1.66 |
| DVU0647 | Iron compound ABC transporter | 1.62 | 0.00 | 0.24 | 0.14 | 1.42 | 0.15 |
| DVU0648 | Iron compound ABC transporter | 1.69 | 0.00 | 0.81 | 0.04 | 1.13 | 0.21 |
| DVU0649 | Iron compound ABC transporter | 0.55 | 5.87 | 0.11 | 0.14 | 0.42 | 3.03 |
| DVU2292 | Hydrogenase nickel insertion protein | −1.38 | 0.00 | −1.10 | 0.00 | −0.31 | 4.88 |
| DVU2402 | Heterodisulfide reductase, A subunit (HdrA) | 1.10 | 0.00 | 0.09 | 0.14 | 1.07 | 0.36 |
| DVU2403 | Heterodisulfide reductase, B subunit (HdrB) | 1.81 | 0.00 | 0.92 | 0.00 | 0.92 | 0.41 |
| DVU2404 | Heterodisulfide reductase, C subunit (HdrC) | 1.59 | 0.00 | 0.57 | 0.04 | 1.09 | 0.93 |
| DVU0587 | Formate dehydrogenase, alpha | 2.95 | 0.00 | 1.61 | 0.00 | 1.45 | 0.00 |
| DVU0588 | Formate dehydrogenase, beta | 2.81 | 0.00 | 1.89 | 0.00 | 2.22 | 0.00 |
| DVU2797 | Cytochrome c553 | 1.23 | 0.00 | 0.69 | 0.00 | 0.56 | 1.80 |
| DVU3171 | Cytochrome c3 (TpI-c3) | −0.65 | 3.59 | −2.45 | 0.00 | 1.86 | 0.00 |
| DVU2201 | Alcohol dehydrogenase | −2.81 | 0.00 | −1.67 | 0.00 | −1.07 | 0.00 |
| DVU2396 | Alcohol dehydrogenase | 1.25 | 0.00 | 0.49 | 0.04 | 0.78 | 1.66 |
| DVU2405 | Alcohol dehydrogenase | −1.30 | 0.06 | 0.06 | 0.19 | −1.43 | 1.66 |
| DVU2545 | Alcohol dehydrogenase | 0.43 | 7.10 | 1.01 | 0.00 | −0.68 | 1.66 |
| DVU2885 | Alcohol dehydrogenase | 0.92 | 0.33 | 1.04 | 0.00 | 0.18 | 5.19 |
| DVU0353 | Alcohol dehydrogenase | −0.49 | 8.60 | 0.63 | 0.11 | −1.13 | 0.36 |
Analysis of gene expression by RT-PCR.
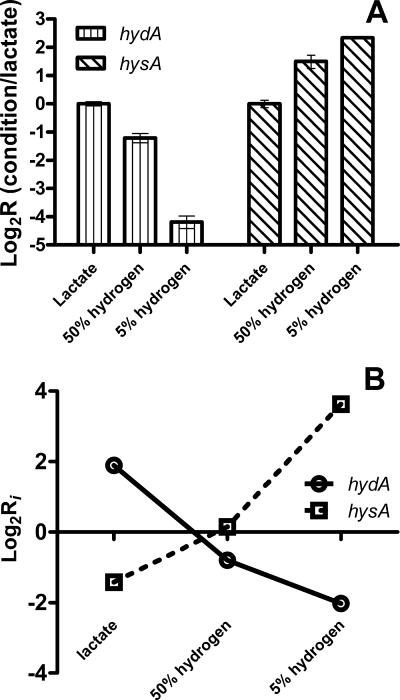
To validate the gene expression changes indicated by the microarray data, RT-PCR was performed on the isolated RNA samples using primers targeting the hysA and hydA genes, encoding the hydrogenase large subunits. The RT-PCR data demonstrated the same trends (Fig. 3A) seen with the microarray data (Fig. 3B). Expression of the hydA gene decreased, whereas expression of the hysA gene increased, when growth conditions changed from lactate to 50% hydrogen and then to 5% hydrogen.
FIG. 3.
Changes in expression levels of hydA and hysA genes. (A) RT-PCR data for each condition (lactate, 50% H2, or 5% H2) were normalized for expression of 16S rRNA and are presented relative to expression with lactate. (B) Microarray hybridization intensity ratios, Ri = <cDNAi>/<gDNAi>, were calculated from measured data as described in the text. The log2Ri values for the hysA and hydA genes are plotted for growth with lactate, 50% (vol/vol) hydrogen, or 5% (vol/vol) hydrogen.
DISCUSSION
Hydrogen not only serves as an energy source for Desulfovibrio spp. in their indigenous environments but also has been implicated in the hydrogen-cycling mechanism for establishing the chemiosmotic gradient required for ATP production during lactate oxidation (18). Hence, growth on lactate may cause a physiological response similar to that of growth on environmental hydrogen. For hydrogen cycling to function, a cytoplasmic hydrogenase is required to generate hydrogen from the protons and electrons liberated during lactate oxidation. The genome sequence of D. vulgaris has indicated the presence of two membrane-bound, cytoplasmically oriented hydrogenases, Coo (DVU2286 to DVU2291) and Ech (DVU0429 to DVU0434). Of these, the genes for Coo hydrogenase appear upregulated during growth on lactate, whereas those for Ech hydrogenase are upregulated during growth on hydrogen (Table 3). Comparing log-phase cultures grown with formate or lactate as the electron donor, Zhang et al. reported similar regulation of Coo hydrogenase genes (37). Consequently, the former may be involved in hydrogen cycling, although it should be pointed out that genes for these membrane-bound, cytoplasmic hydrogenases are absent from the closely related D. desulfuricans G20 genome, and hence a mechanism involving them cannot have general validity. Growth on hydrogen upregulated one of three periplasmic formate dehydrogenases (DVU0587 and DVU0588) (Table 3), which may function in the direction of formate synthesis (CO2 + H+ + 2e → HCOO−) under these conditions.
Studies with D. vulgaris and other microorganisms have established that hydrogenases are regulated by a number of different environmental signals. Effectors include the availability of iron (12), nickel (33), selenium (30), and electron acceptors such as oxygen (8, 19). We have shown that the electron donor used, either lactate or hydrogen, as well as the concentration of hydrogen, also strongly regulates hydrogenase gene expression in D. vulgaris. Our results generally agree with those of a study using rich medium (1 g/liter of yeast extract) in which the growth of D. vulgaris with lactate was compared to that with hydrogen (80% vol/vol) as the electron donor. The hys genes were found to be upregulated, whereas the hyd genes were unchanged in that study (20). Additionally, we have shown that the elimination of a particular hydrogenase can impact growth differently as the environmental hydrogen concentration is altered. Correlation of the gene expression and growth data indicates that when the hydrogen partial pressure is decreased, the [Fe] hydrogenase becomes less important, while the [NiFeSe] hydrogenase becomes more important, to the organism's fitness. This relationship complements the kinetic properties of these two hydrogenases detailed in the introduction: the [Fe] hydrogenase has a higher specific activity but lower binding affinity, making it more efficient at higher hydrogen concentrations (7), while the [NiFeSe] hydrogenase has a higher binding affinity but a lower specific activity, allowing it to operate more efficiently at lower hydrogen concentrations (23, 31, 32). Although details of the hydrogen-cycling model of lactate utilization by Desulfovibrio spp. remain unclear, lactate oxidation may be regarded as a special case of growth on hydrogen, in which hydrogen is derived from lactate oxidation rather than from the environment. If the expression levels from either the microarray or RT-PCR data for growth with 50 and 5% hydrogen are extrapolated to include growth with lactate, it appears that the latter condition corresponds to a concentration of hydrogen greater than 50%. The expression of the hyd gene increases from 5% hydrogen to 50% hydrogen to lactate, whereas that of the hys gene decreases as the growth medium is changed from 5% hydrogen to 50% hydrogen to lactate (Fig. 3).
The expression of D. vulgaris hydrogenases appears to be regulated by metal availability at the protein level (30). Activity stain and protein blotting experiments showed that [Fe] hydrogenase is present at the highest concentrations when the medium contains only added iron (25 μM), that [NiFe] hydrogenase 1 is somewhat induced by additional nickel (1 μM), and that expression of [NiFeSe] hydrogenase strictly depends on addition of 1 μM selenium to the medium, in addition to 25 μM iron and 1 μM nickel. The [NiFeSe] hydrogenase was found to be the dominant hydrogenase under these conditions (30). At the transcription level, such strong effects were not observed: addition of nickel downregulated hydA and upregulated hynA1 somewhat, whereas addition of selenium hardly upregulated the hys genes (30). We added less selenium and nickel (22 nM and 100 nM, respectively, the normal concentrations in WP medium) and found that, under these conditions, the level of the hys message is strongly regulated by the hydrogen concentration (Fig. 3). Hydrogen may exert master regulation over expression of genes for hydrogenases through the NikR regulator. Although hydrogen exposure did not change the expression level of the nikR gene, the nickel transport operon nikOQMK, which usually is under the control of nikR, was upregulated when D. vulgaris was grown with hydrogen (Table 3). Similarly, the iron transport system DVU0647 to DVU0649 was upregulated with hydrogen exposure.
In addition to the hydrogenases, D. vulgaris also produces a variety of soluble cytochrome c3's and transmembrane electron transport complexes (14). It has been suggested that the multiple periplasmic hydrogenases feed protons and electrons through distinct networks, represented by these cytochromes and transmembrane electron transport complexes, that ultimately reach the cytoplasmic sulfate reduction enzymes (13, 14). The most well-known cytochrome c3, TpI-c3 (DVU3171), was downregulated with 5% hydrogen, relative to growth with lactate and with 50% hydrogen, suggesting that it may be more important as a physiological electron donor to [Fe] hydrogenase than to [NiFeSe] hydrogenase, although in vitro it has been shown to work with both (31). Genes for the Hmc complex (DVU0531 to DVU0536) appeared somewhat downregulated during growth with hydrogen (Table 3), despite earlier findings that an hmc mutant has partially impaired growth with hydrogen (4).
Although it is clear that hydrogen concentration affects the expression of the [NiFeSe] and [Fe] hydrogenase genes, it did not display a clear effect on the expression of the [NiFe] hydrogenase 1 or [NiFe] hydrogenases 2 isozymes. It should be noted that although deletion of the hys and hyd genes affected growth at low and high hydrogen concentration, respectively, all mutants grew to some extent in all conditions (Fig. 2). This indicates that the remaining two [NiFe] hydrogenase isozymes are able to compensate for the absence of either the [NiFeSe] or [Fe] hydrogenase, and that these overlapping functions are advantageous and may provide a rationale for the hydrogenase redundancy. The four periplasmic hydrogenases also differ in their susceptibilities to various inhibitors. The [Fe] hydrogenase is more sensitive to CO, NO, and NO2− inhibition than the other hydrogenases, and the [NiFeSe] hydrogenase is more sensitive to CO inhibition than [NiFe] hydrogenase 1 (7). This difference in inhibition sensitivity may provide another rationale for the existence of additional nickel hydrogenase enzymes, and this possibility needs to be explored in more detail.
Supplementary Material
Acknowledgments
This work was supported by a Strategic Grant and a Discovery Grant from the Natural Science and Engineering Research Council (NSERC) of Canada to G.V. The microarray analysis was supported by the U.S. Department of Energy under the Genomics:GTL program through the Virtual Institute of Microbial Stress and Survival (VIMSS; http://vimss.lbl.gov), the Office of Biological and Environmental Research, Office of Science.
We thank Judy Wall for sharing how to use G418 for mutagenesis of D. vulgaris. Ines Pereira and Ricardo Louro are thanked for making the Ph.D. thesis of Patricia Pereira (20) available following the completion of this work.
Footnotes
Published ahead of print on 29 June 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexeyev, M. F., and I. N. Shokolenko. 1995. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of gram negative bacteria. Gene 160:59-62. [DOI] [PubMed] [Google Scholar]
- 2.Bender, K. S., H.-C. Yen, and J. D. Wall. 2006. Analysing the metabolic capabilities of Desulfovibrio species through genetic manipulation. Biotechnol. Genet. Eng. Rev. 23:157-174. [DOI] [PubMed] [Google Scholar]
- 3.Chhabra, S. R., Q. He, K. H. Huang, S. P. Gaucher, E. J. Alm, Z. He, M. Z. Hadi, T. C. Hazen, J. D. Wall, J. Zhou, A. P. Arkin, and A. K. Singh. 2006. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188:1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolla, A., B. K. Pohorelic, J. K. Voordouw, and G. Voordouw. 2000. Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch. Microbiol. 174:143-151. [DOI] [PubMed] [Google Scholar]
- 5.Edgar, R. M., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst, F. D., S. Bereswill, B. Waidner, J. Stoof, U. Mader, J. G. Kusters, E. J. Kuipers, M. Kist, A. H. van Vliet, and G. Homuth. 2005. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151:533-546. [DOI] [PubMed] [Google Scholar]
- 7.Fauque, G., H. D. Peck, Jr., J. J. Moura, B. H. Huynh, Y. Berlier, D. V. DerVartanian, M. Teixeira, A. E. Przybyla, P. A. Lespinat, and I. Moura. 1988. The three classes of hydrogenases from sulfate-reducing bacteria of the genus Desulfovibrio. FEMS Microbiol. Rev. 4:299-344. [DOI] [PubMed] [Google Scholar]
- 8.Fournier, M., Z. Dermoun, M. C. Durand, and A. Dolla. 2004. A new function of the Desulfovibrio vulgaris Hildenborough [Fe] hydrogenase in the protection against oxidative stress. J. Biol. Chem. 279:1787-1793. [DOI] [PubMed] [Google Scholar]
- 9.Frey, M. 2002. Hydrogenases: hydrogen-activating enzymes. Chembiochem 3:153-160. [DOI] [PubMed] [Google Scholar]
- 10.Fu, R., and G. Voordouw. 1997. Targeted gene-replacement mutagenesis of dcrA, encoding an oxygen sensor of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Microbiology 143:1815-1826. [DOI] [PubMed] [Google Scholar]
- 11.Goenka, A., J. K. Voordouw, W. Lubitz, W. Gaertner, and G. Voordouw. 2005. Construction of a [NiFe]-hydrogenase deletion mutant of Desulfovibrio vulgaris Hildenborough. Biochem. Soc. Trans. 33:59-60. [DOI] [PubMed] [Google Scholar]
- 12.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 13.Haveman, S. A., E. A. Greene, and G. Voordouw. 2005. Gene expression analysis of the mechanism of inhibition of Desulfovibrio vulgaris Hildenborough by nitrate-reducing, sulfide-oxidizing bacteria. Environ. Microbiol. 7:1461-1465. [DOI] [PubMed] [Google Scholar]
- 14.Heidelberg, J. F., R. Seshadri, S. A. Haveman, C. L. Hemme, I. T. Paulsen, J. F. Kolonay, J. A. Eisen, N. Ward, B. Methe, L. M. Brinkac, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, D. Fouts, D. H. Haft, J. Selengut, J. D. Peterson, T. M. Davidsen, N. Zafar, L. Zhou, D. Radune, G. Dimitrov, M. Hance, K. Tran, H. Khouri, J. Gill, T. R. Utterback, T. V. Feldblyum, J. D. Wall, G. Voordouw, and C. M. Fraser. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22:554-559. [DOI] [PubMed] [Google Scholar]
- 15.Hubert, C., and G. Voordouw. 2007. Oil field souring control by nitrate-reducing Sulfurospirillum spp. that outcompete sulfate-reducing bacteria for organic electron donors. Appl. Environ. Microbiol. 73:2644-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huynh, B. H., M. H. Czechowski, H. J. Kruger, D. V. DerVartanian, H. D. Peck, Jr., and J. LeGall. 1984. Desulfovibrio vulgaris hydrogenase: a nonheme iron enzyme lacking nickel that exhibits anomalous EPR and Mossbauer spectra. Proc. Natl. Acad. Sci. USA 81:3728-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matias, P. M., I. A. Pereira, C. M. Soares, and M. A. Carrondo. 2005. Sulphate respiration from hydrogen in Desulfovibrio bacteria: a structural biology overview. Prog. Biophys. Mol. Biol. 89:292-329. [DOI] [PubMed] [Google Scholar]
- 18.Odom, J. M., and H. D. Peck, Jr. 1984. Hydrogenase, electron-transfer proteins, and energy coupling in the sulfate-reducing bacteria Desulfovibrio. Annu. Rev. Microbiol. 38:551-592. [DOI] [PubMed] [Google Scholar]
- 19.Olson, J. W., and R. J. Maier. 2002. Molecular hydrogen as an energy source for Helicobacter pylori. Science 298:1788-1790. [DOI] [PubMed] [Google Scholar]
- 20.Pereira, P. M. 2006. Biochemical and genomic studies of proteins involved in the bioenergetic metabolism of sulfate-reducing bacteria. Ph.D. thesis. Instituto de Tecnologiaq Uimica e Biologica, Universidade Nova de Lisboa, Lisbon, Spain.
- 21.Pohorelic, B. K., J. K. Voordouw, E. Lojou, A. Dolla, J. Harder, and G. Voordouw. 2002. Effects of deletion of genes encoding Fe-only hydrogenase of Desulfovibrio vulgaris Hildenborough on hydrogen and lactate metabolism. J. Bacteriol. 184:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postgate, J. R. 1984. The sulphate-reducing bacteria, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 23.Romão, C. V., I. A. Pereira, A. V. Xavier, J. LeGall, and M. Teixeira. 1997. Characterization of the [NiFe] hydrogenase from the sulfate reducer Desulfovibrio vulgaris Hildenborough. Biochem. Biophys. Res. Commun. 240:75-79. [DOI] [PubMed] [Google Scholar]
- 24.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 25.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 26.Simon, R., U. Priefer, and A. Pühler. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 27.Talaat, A. M., S. T. Howard, W. T. Hale, R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 29.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valente, F. M., C. C. Almeida, I. Pacheco, J. Carita, L. M. Saraiva, and I. A. Pereira. 2006. Selenium is involved in regulation of periplasmic hydrogenase gene expression in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188:3228-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valente, F. M., A. S. Oliveira, N. Gnadt, I. Pacheco, A. V. Coelho, A. V. Xavier, M. Teixeira, C. M. Soares, and I. A. Pereira. 2005. Hydrogenases in Desulfovibrio vulgaris Hildenborough: structural and physiologic characterisation of the membrane-bound [NiFeSe] hydrogenase. J. Biol. Inorg. Chem. 10:667-682. [DOI] [PubMed] [Google Scholar]
- 32.van Haaster, D. J., P. L. Hagedoorn, J. A. Jongejan, and W. R. Hagen. 2005. On the relationship between affinity for molecular hydrogen and the physiological directionality of hydrogenases. Biochem. Soc. Trans. 33:12-14. [DOI] [PubMed] [Google Scholar]
- 33.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 34.Voordouw, G. 2002. Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 184:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voordouw, G., V. Niviere, F. G. Ferris, P. M. Fedorak, and D. W. Westlake. 1990. Distribution of hydrogenase genes in Desulfovibrio spp. and their use in identification of species from the oil field environment. Appl. Environ. Microbiol. 56:3748-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3353-3378. In H. G. T. A. Balows, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer, New York, NY.
- 37.Zhang, W., D. E. Culley, J. C. Scholten, M. Hogan, L. Vitiritti, and F. J. Brockman. 2006. Global transcriptomic analysis of Desulfovibrio vulgaris on different electron donors. Antonie Leeuwenhoek 89:221-237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.