Abstract
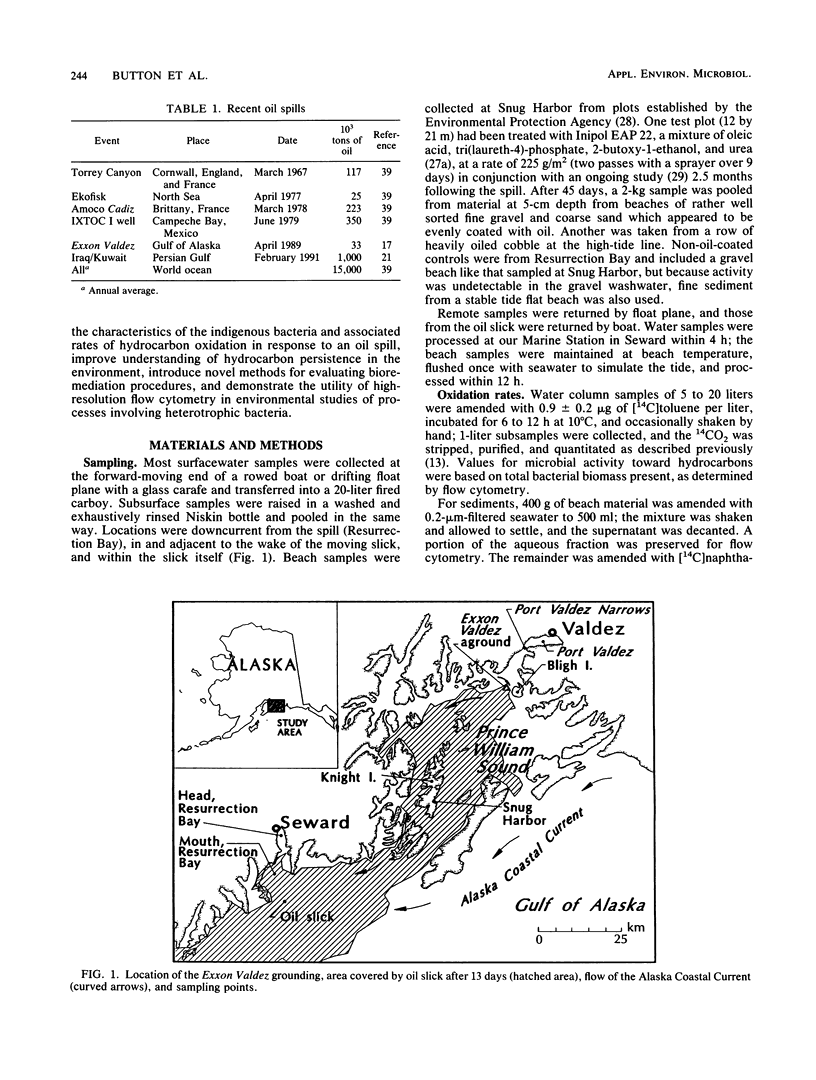
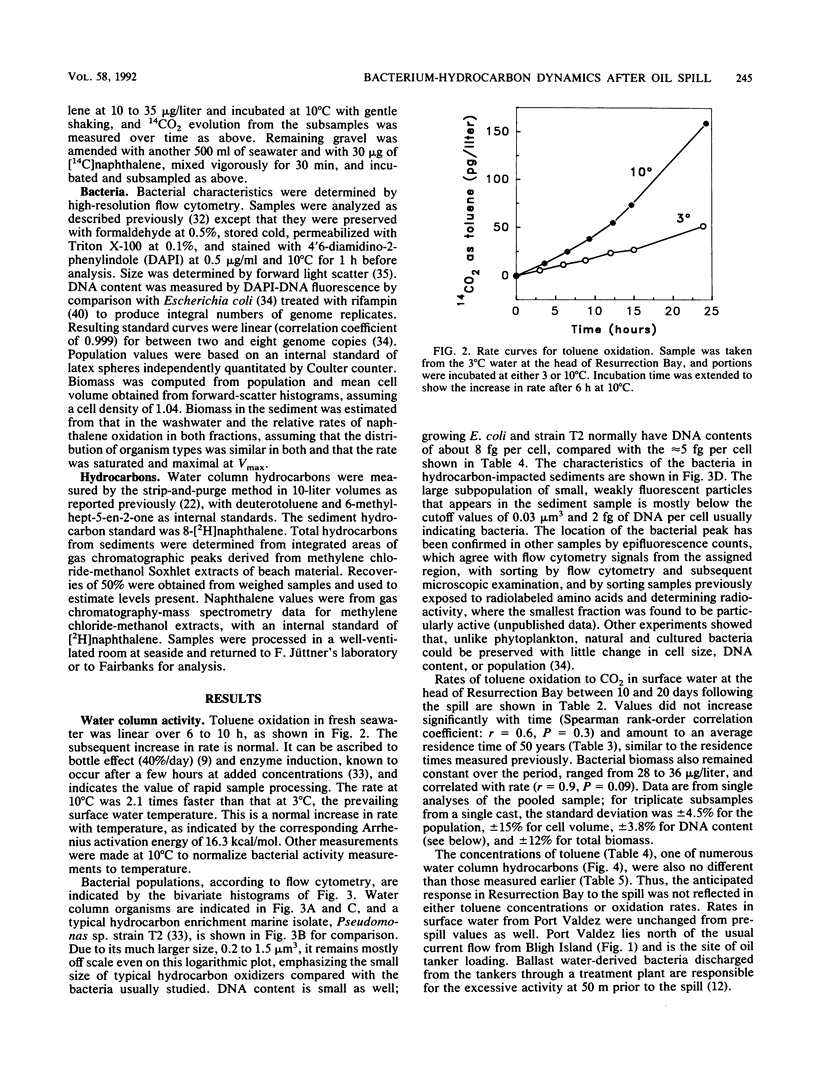
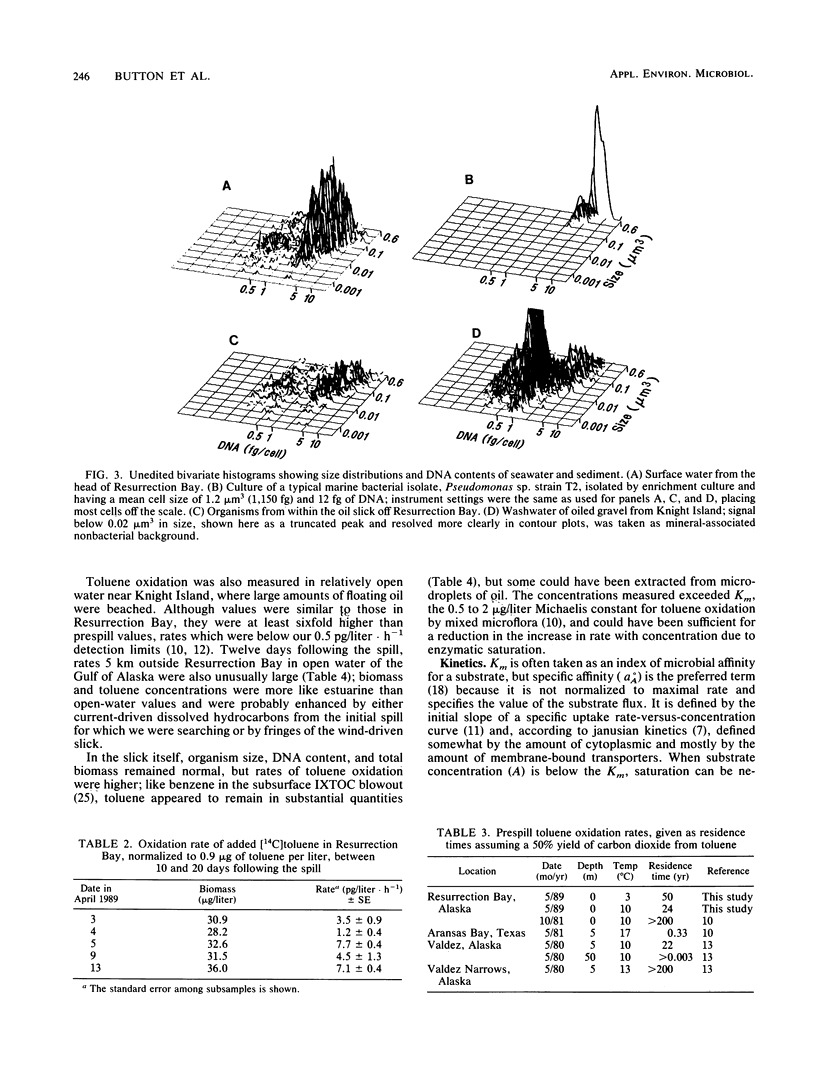
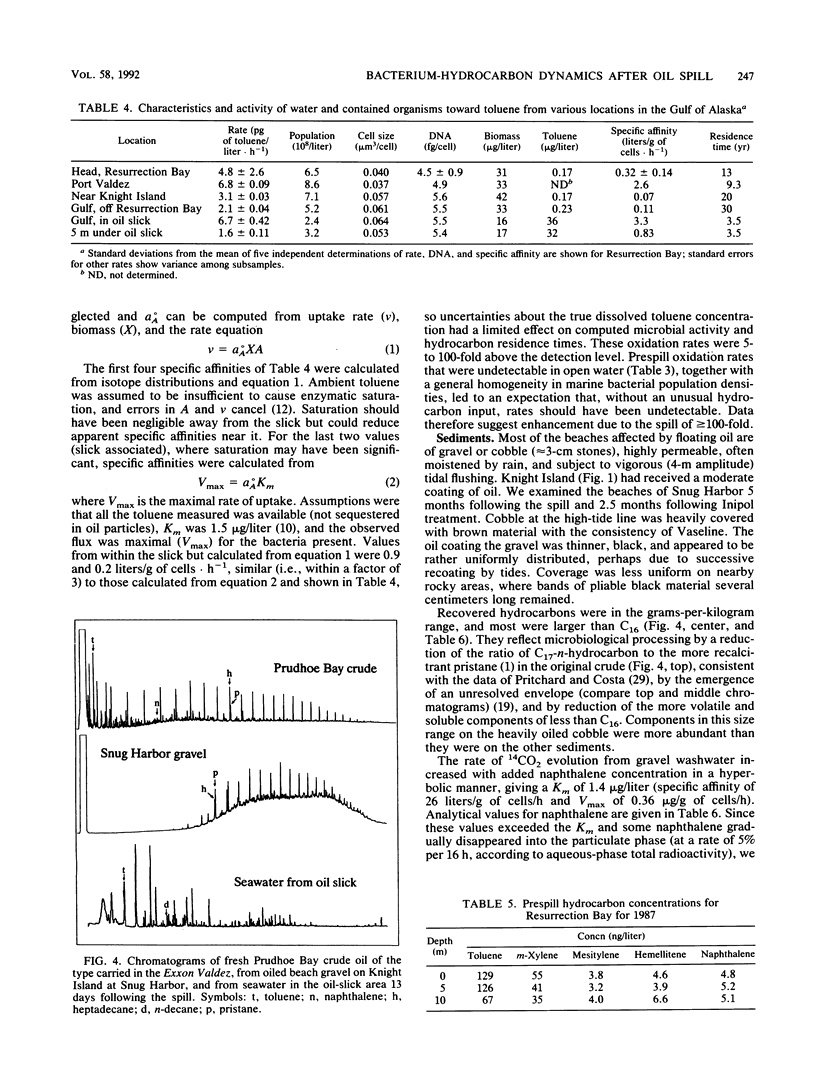
Turnover times for toluene in Resurrection Bay after the Exxon Valdez grounding were determined to be decades, longer than expected considering that dissolved hydrocarbons were anticipated to drift with the current and stimulate development of additional hydrocarbon-utilizing capacity among the microflora in that downcurrent location. These turnover times were based on the recovery of 14CO2 from added [14C]toluene that was oxidized. The concentrations of toluene there, 0.1 to 0.2 microgram/liter, were similar to prespill values. Oxidation rates appeared to be enhanced upstream near islands in the wake of the wind-blown slick, and even more within the slick itself. Specific affinities of the water column bacteria for toluene were computed with the help of biomass data, as measured by high-resolution flow cytometry. They were a very low 0.3 to 3 liters/g of cells.h-1, indicating limited capacity to utilize this hydrocarbon. Since current-driven mixing rates exceeded those of oxidation, dissolved spill components such as toluene should enter the world-ocean pool of hydrocarbons rather than biooxidize in place. Some of the floating oil slick washed ashore and permeated a coarse gravel beach. A bacterial biomass of 2 to 14 mg/kg appeared in apparent response to the new carbon and energy source. This biomass was computed from that of the organisms and associated naphthalene oxidation activity washed from the gravel compared with the original suspension. These sediment organisms were very small at approximately 0.06 microns 3 in volume, low in DNA at approximately 5.5 g per cell, and unlike the aquatic bacteria obtained by enrichment culture but quite similar to the oligobacteria in the water column.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aelion C. M., Bradley P. M. Aerobic biodegradation potential of subsurface microorganisms from a jet fuel-contaminated aquifer. Appl Environ Microbiol. 1991 Jan;57(1):57–63. doi: 10.1128/aem.57.1.57-63.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M. Biodegradation of chemicals of environmental concern. Science. 1981 Jan 9;211(4478):132–138. doi: 10.1126/science.7444456. [DOI] [PubMed] [Google Scholar]
- Button D. K. Biochemical basis for whole-cell uptake kinetics: specific affinity, oligotrophic capacity, and the meaning of the michaelis constant. Appl Environ Microbiol. 1991 Jul;57(7):2033–2038. doi: 10.1128/aem.57.7.2033-2038.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button D. K. Evidence for a terpene-based food chain in the gulf of alaska. Appl Environ Microbiol. 1984 Nov;48(5):1004–1011. doi: 10.1128/aem.48.5.1004-1011.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button D. K. Kinetics of nutrient-limited transport and microbial growth. Microbiol Rev. 1985 Sep;49(3):270–297. doi: 10.1128/mr.49.3.270-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button D. K., Robertson B. R., Craig K. S. Dissolved hydrocarbons and related microflora in a fjordal seaport: sources, sinks, concentrations, and kinetics. Appl Environ Microbiol. 1981 Oct;42(4):708–719. doi: 10.1128/aem.42.4.708-719.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button D. K., Robertson B. R. Kinetics of bacterial processes in natural aquatic systems based on biomass as determined by high-resolution flow cytometry. Cytometry. 1989 Sep;10(5):558–563. doi: 10.1002/cyto.990100511. [DOI] [PubMed] [Google Scholar]
- Button D. K., Schell D. M., Robertson B. R. Sensitive and accurate methodology for measuring the kinetics of concentration-dependent hydrocarbon metabolism rates in seawater by microbial communities. Appl Environ Microbiol. 1981 Apr;41(4):936–941. doi: 10.1128/aem.41.4.936-941.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford M. Bacteria effective in Alaska cleanup. Science. 1990 Mar 30;247(4950):1537–1537. doi: 10.1126/science.2321011. [DOI] [PubMed] [Google Scholar]
- Donachie W. D., Begg K. J. Cell length, nucleoid separation, and cell division of rod-shaped and spherical cells of Escherichia coli. J Bacteriol. 1989 Sep;171(9):4633–4639. doi: 10.1128/jb.171.9.4633-4639.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschal J. C. Some reflections on microbial competitiveness among heterotrophic bacteria. Antonie Van Leeuwenhoek. 1985;51(5-6):473–494. doi: 10.1007/BF00404494. [DOI] [PubMed] [Google Scholar]
- Powers D. A. Fish as model systems. Science. 1989 Oct 20;246(4928):352–358. doi: 10.1126/science.2678474. [DOI] [PubMed] [Google Scholar]
- Robertson B. R., Button D. K. Characterizing aquatic bacteria according to population, cell size, and apparent DNA content by flow cytometry. Cytometry. 1989 Jan;10(1):70–76. doi: 10.1002/cyto.990100112. [DOI] [PubMed] [Google Scholar]
- Robertson B. R., Button D. K. Phosphate-limited continuous culture of Rhodotorula rubra: kinetics of transport, leakage, and growth. J Bacteriol. 1979 Jun;138(3):884–895. doi: 10.1128/jb.138.3.884-895.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson B. R., Button D. K. Toluene induction and uptake kinetics and their inclusion in the specific-affinity relationship for describing rates of hydrocarbon metabolism. Appl Environ Microbiol. 1987 Sep;53(9):2193–2205. doi: 10.1128/aem.53.9.2193-2205.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubal G., Atlas R. M. Distribution of hydrocarbon-utilizing microorganisms and hydrocarbon biodegradation potentials in Alaskan continental shelf areas. Appl Environ Microbiol. 1978 May;35(5):897–905. doi: 10.1128/aem.35.5.897-905.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen H. B., Boye E. Escherichia coli growth studied by dual-parameter flow cytophotometry. J Bacteriol. 1981 Feb;145(2):1091–1094. doi: 10.1128/jb.145.2.1091-1094.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylstra G. J., McCombie W. R., Gibson D. T., Finette B. A. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol. 1988 Jun;54(6):1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]


