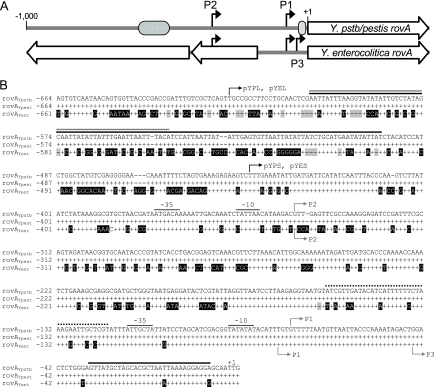
FIG. 1.
rovA promoters of Yersinia. (A) Linear representation of comparisons of the rovA regions from Y. pseudotuberculosis/Y. pestis (Y.pstb/pestis rovA) and Y. enterocolitica (Y. enterocolitica rovA). The black arrows indicate transcriptional initiation sites. The cross-hatched oval represents the predicted high-affinity RovA/H-NS binding site. The gray oval represents the predicted low-affinity RovA binding site. The large open arrows represent ORFs. (B) Sequence alignment of DNA upstream of rovA in Y. pseudotuberculosis (rovAYpstb) with similar regions in Y. pestis (rovAYpest) and Y. enterocolitica (rovAYent). Conserved nucleotides are represented by plus signs, and divergent nucleotides are indicated by a black background. Gaps are indicated by shaded dashes. The initiation codon for rovA is designated +1. The gray arrows indicate mRNA initiation sites for Y. pseudotuberculosis (above the sequences) or Y. enterocolitica (below the sequences). Predicted −35 and −10 regions for P1 and P2 in Y. pseudotuberculosis are also shown. Solid black, gray, and dotted lines above the sequences indicate predicted RovA, H-NS, and RovM binding sites, respectively, in Y. pseudotuberculosis (17, 18). The black arrows indicate the locations of the most 5′ nucleotides of reporter constructs.