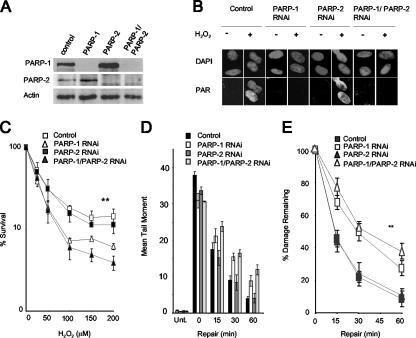
FIG. 2.
Depletion of PARP-1 but not PARP-2 reduces rates of chromosomal SSBR and sensitizes human A549 cells to oxidative DNA damage. (A) Levels of PARP-1 and PARP-2 protein in total cells extracts from A549 cells transfected with pcD2E and either empty pSuper (Control), pSuper-PARP-1 (PARP-1), pSuper-PARP-2 (PARP-2), or both pSuper-PARP-1 and pSuper-PARP-2 (PARP-1/PARP-2), as measured by immunoblotting with appropriate antibodies. (B) Levels of PAR before and after (1-min repair) treatment with 10 mM H2O2 on ice with A549 cells depleted of the indicated proteins, as measured by indirect immunofluorescence microscopy. Cells were counterstained with DAPI to identify nuclear DNA. (C) Clonogenic survival of A549 cells depleted of the indicated proteins following exposure to the indicated concentrations of H2O2 in PBS for 10 min at RT. Cells were fixed after 14 days and stained with methylene blue, and the fraction (%) of surviving cells was calculated. Data are the means (± 1 SE) of three independent experiments. **, the survival curve for control cells was significantly different (by analysis of variance [ANOVA]) from those of PARP-1-depleted (P = 0.009) and PARP-1/PARP-2-depleted (P = 0.01) cells but not those of PARP-2-depleted cells (P = 0.57). The survival curves for PARP-1- and PARP-1/PARP-2-depleted cells were not significantly different (P = 0.76). (D) Total DNA strand breakage was quantified by comet assays with A549 cells depleted of the indicated proteins before (Unt.) and immediately after (0) treatment with 100 μM H2O2 for 20 min on ice and after the indicated repair periods in H2O2-free medium. Data points are the means (± 1 SE) of at least three independent experiments, with the average tail moment from 100 cells calculated in each experiment. (E) The data from panel D were replotted as the fraction (%) of DNA strand breaks remaining at the indicated DNA repair time points. **, the repair kinetics for control cells were statistically significantly (ANOVA) different from those of PARP-1-depleted (P = 7.4 × 10−7) and PARP-1/PARP-2-depleted (P = 0.68 × 10−8) cells but not those of PARP-2-depleted cells (P = 0.87). The repair kinetics for PARP-1- and PARP-1/PARP-2-depleted cells were not significantly different (P = 0.1).