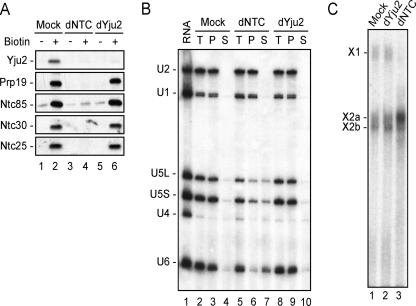
FIG. 5.
Yju2 was not required for spliceosome activation. (A) Splicing reactions were carried out in mock-depleted (lanes 1 and 2), NTC-depleted (lanes 3 and 4), or Yju2-depleted (lanes 5 and 6) extract using biotinylated (lanes 2, 4, and 6) or nonbiotinylated (lanes 1, 3, and 5) ACAC pre-mRNA. The spliceosome was isolated by precipitation with streptavidin-Sepharose, followed by Western blotting using antibody against Yju2, Prp19, Ntc85, Ntc30, or Ntc25. (B) Splicing reactions were carried out in mock-depleted (lanes 2 to 4), NTC-depleted (lanes 5 to 7), or Yju2-depleted (lanes 8 to 10) extract using biotinylated ACAC pre-mRNA as the substrate, and the spliceosome was precipitated with streptavidin-Sepharose. After the unbound materials were washed off, the pellet was separated into two fractions: one was the total precipitate (T; lanes 2, 5, and 8), and the other was added to the splicing buffer and incubated at room temperature for 20 min. After separating the supernatant (S; lanes 4, 7, and 10) and pellet fractions (P; lanes 3, 6, and 9), RNA was extracted and analyzed by Northern blotting. Lane 1 is total RNA from 2 μl of extracts. (C) Splicing reactions were carried out in mock-depleted (lane 1), Yju2-depleted (lane 2), or NTC-depleted extract (lane 3) using Ac/Cla pre-mRNA as the substrate. The reaction mixtures were precipitated with the anti-Smd1 antibody, followed by UV irradiation. RNA was then isolated and selected with 5′-end-biotinylated oligonucleotide U6-Abio through precipitation with streptavidin-Sepharose and was then further analyzed. The cross-linked products X1, X2a, and X2b are as described by Chan and Cheng (5). dYju2, Yju2-depleted; dNTC, NTC-depleted.