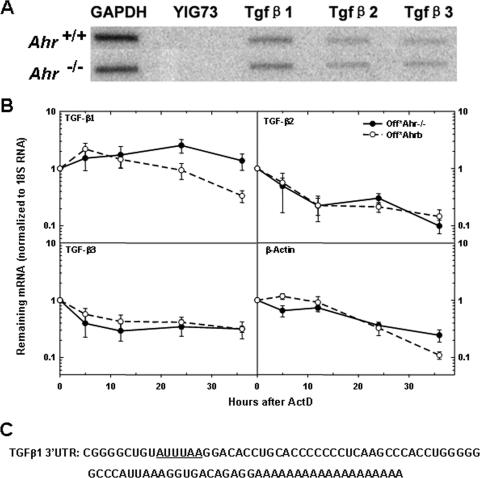
FIG. 5.
AHR destabilizes TGF-β1 mRNA. (A) Nuclear run-on assays of Ahr−/− MEFs and wild-type MEFs. Nuclear run-on assays were conducted as described in Materials and Methods. The hybridization targets were the PCR products of TGF-β1, TGF-β2, and TGF-β3. YIG73 is a yeast intergenic region used as negative control, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a positive control. (B) Analysis of mRNA stability. Quiescent Off*Ahr−/− and Off*Ahrb cells were stimulated to enter the cell cycle with MEM-α containing 10% serum for 40 h and further incubated with actinomycin D (ActD) at a final concentration of 0.05 μg/ml for 36 h. mRNA levels at the various time intervals were determined by quantitative real-time RT-PCR. The results are expressed as the percentage of mRNA remaining relative to the corresponding level before the addition of ActD. Data are the means ± standard deviations (error bars) of three independent measurements, each with technical triplicates of duplicate biological samples for each cell type at each time point. (C) Sequence of 3′UTR of TGF-β1 mRNA, with AUUUAA region underlined.