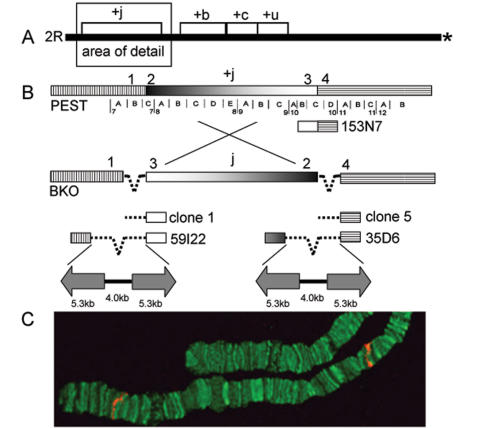
Figure 2. Cloning strategy for the 2Rj inversion breakpoints.
(A) Relative position of four paracentric inversions (j, b, c, u) on chromosome 2R in An. gambiae. Asterisk represents the centromere. (B) Schematic of alternative arrangements of 2Rj in PEST (+j) and BKO (j) strains (not to scale). The cytogenetic map represented beneath the PEST chromosome shows the breakpoints positioned in subdivision 7C and 10C. Numbers above the chromosomes are used in the text to refer to corresponding regions adjacent to the breakpoints, represented as identically hatched or shaded regions in alternative arrangements. Dashed line indicates repetitive DNA insertion. Shown beneath the PEST chromosome is a representation of BAC 153N7 that spans the proximal breakpoint. Below the BKO chromosome are plasmid (1, 5) and fosmid (59I22, 35D6) clones corresponding to the breakpoints of the j arrangement. Structure of the repetitive DNA in 59I22 and 35D6 is represented below as arrows (inverted repeats) separated by a solid line (central spacer) of indicated lengths in kb. (C) FISH performed on the chromosomes of the BKO strain (2Rj) with BAC 153N7. Signals in red indicate the distal and the proximal breakpoints of the j inversion.